Influenza A virus (IAV) infections pose a major challenge in research and practice worldwide, not only in sows but also in piglets and fatteners. Influenza viruses are now endemic on many pig farms. The clinical picture is widely variable. Diagnosis is often complicated by co-infections and, in many cases, pathogen detection beyond the clinically affected age groups is required to identify the influenza virus involved in the disease process. The increasing dynamic in the genetic evolution of influenza viruses and the variation in infectivity and disease expression require experience and knowledge for interpreting the diagnostic results. In this first part of three consecutive articles, we focus on latest news about the virus and the diagnostic approach. The second part details clinical disease and co-infections and the third part gives an overview about control strategies.
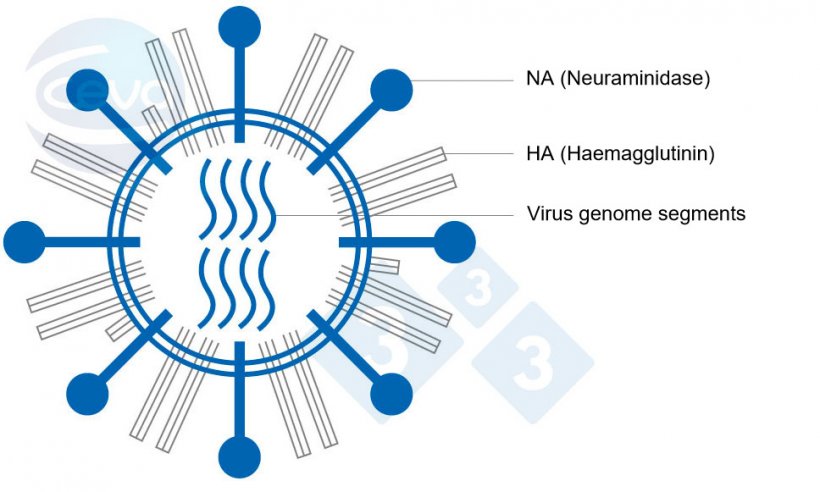
Influenza A (IAV) viruses in pigs are enveloped RNA viruses belonging to the family of Orthomyxoviridae. A characteristic feature is the genome consisting of eight gene segments, each coding at least for one protein. Two segments among the eight are responsible for the surface antigenic structures haemagglutinin (HA) and neuraminidase (NA) (Fig. 1). IAV are classified in subtypes according to these structures. Eighteen HA and eleven NA subtypes have been identified to date.

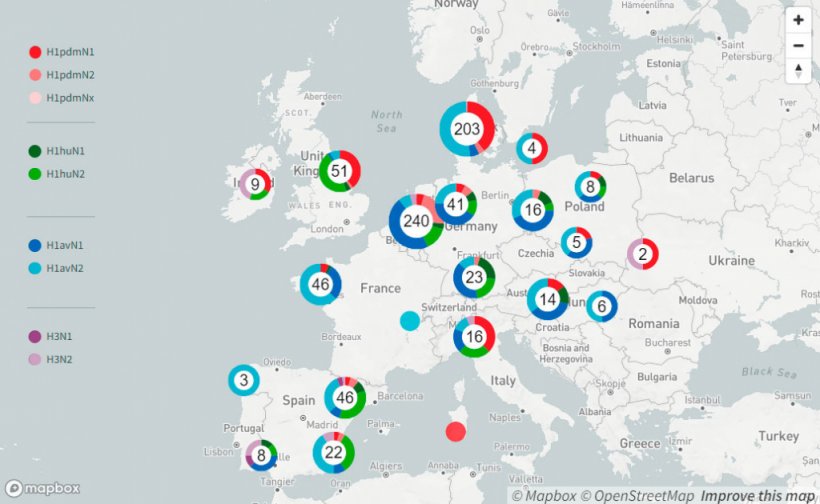
Until 2009, there were mainly three Influenza A virus subtypes circulating in swine (swIAV), H1avN1, H1huN2 and H3huN2. At that point of time only a few different genotypes were identified, which are defined by all genome segments. This scenario changed with the introduction of the pandemic strain H1pdmN1pdm during the Influenza pandemic in 2009. Recently published data (Henritzi et al. 2020) show that nowadays 31 different swIAV genotypes resulting from reassortment of all eight genome segments between H1pdmN1pdm and the older swIAV lineages are currently circulating in Europe. This passive surveillance was conducted on 2.457 farms in seventeen European countries from 2015 to 2017. It revealed that swIAV can be detected on farms throughout the year and therefore is not seasonal, unlike human influenza. Differences were found in the distribution of subtypes in the various regions.
A range of diagnostic methods are available for the direct detection of swIAV. Methods and number of samples should be chosen depending on the herd situation. Nasal swabs are suitable for influenza A matrix PCR. If positive with a suitable viral load, typing can be conducted by HA- and NA-specific RT-PCR. It should be noted that animals generally excrete the virus for only five to seven days after infection and eliminate it from the lungs after about nine days. Therefore, the timepoint of sampling by individual methods (nasal swabs, broncho-alveolar lavage, lungs), is essential. Since the clinical picture is often complicated by secondary infections, swIAV may no longer be detectable when the most severe symptoms occur. In case of infections with multiple swIAV subtypes, not all subtypes involved may be equally well detected. Especially in farms with endemic influenza and unclear clinical symptoms, it is recommended to use screening methods (udder swab samples, oral fluid samples) in different age groups.

If five nasal swabs in a pooled sample are tested jointly by PCR, there is virtually no loss of sensitivity. The detection rate can be increased because with pools more samples can be processed with the same effort. The nasal swab therefore remains crucial for influenza diagnosis. The good sample quality often allows typing, and culture. It is important that samples are sent to the laboratory cooled by express delivery. In an acute clinical outbreak, samples should be taken from newly sick animals with fever. In endemically infected herds, the selection of animals for sampling should be adjusted to the fact that animals with typical symptoms such as high temperature are less apparent. Virus detection is often possible early in suckling piglets, without clinical signs being visible. As maternal antibodies protect suckling piglets against disease, not against infection, suckling piglets can be shedders and carry the virus into the nursery. Due to mixing of piglets enabling the virus to spread very quickly, the period shortly after weaning is a good timepoint for sampling. Also animals in age groups that regularly show clinical signs (sneezing, sniffling, coughing, fever, suddenly falling behind) should be sampled. Thus, cross-sectional testing is favourable, sampling different age groups at the same timepoint to get the best overview of the circulating swIAV strains on farm.
As an indirect method (detection of antibodies), a "haemagglutination inhibition (HI) test" on serum samples from preferably unvaccinated animals can indicate other subtypes involved that were not previously detected directly. The HI is based on the fact that the blood-clotting effect of IAV is inhibited by antibodies. If a pig´s serum can prevent blood-clotting by the Influenza virus strains used in the test, this is an indication that the pig has developed specific antibodies against similar subtypes. Thus, the HI assay offers an advantage over the ELISA which can only give the information on Influenza A in general. However, serum samples from piglets up to the end of the nursery stage often don’t show antibodies, even if the animals had contact to swIAV in nursery. Seroconversion can be prevented by maternal derived antibodies, passed on to the piglets not only by vaccinated but also by previously infected sows. Therefore, it is not conclusive to blood sample piglets in nursery, however paired serum samples of fatteners or gilts/ sows can provide good results.
Although diagnostics are often very laborious, they are the only way to determine the strains circulating on the farm and the timing of infection. This is a prerequisite for implementing the right management measures and vaccination protocols.