Diarrhoea is one of the major causes of neonatal mortality, decreased weight gain and high use of medication in pigs. The main pathogens are bacteria like Escherichia coli (E. coli) and Clostridium perfringens (C. perfringens) as well as Rota- and Coronaviruses.
A study using samples from a monitoring program in different European countries evaluated the occurrence of these pathogens in the faeces of piglets suffering from neonatal diarrhoea in 2020.

Material and Methods
All samples used in this study derived from suckling piglets. A total of 324 mostly faecal samples from 116 farms were examined bacteriologically for E. coli, C. perfringens and Clostridioides difficile (C. difficile). Samples from 79 of these farms were additionally examined via PCR for Rotavirus group A and C as well as for Porcine Epidemic Virus (PEDV) and Transmissible Gastroenteritis Virus (TGEV). The study therefore consisted of two analyses, based on this difference in the pathogen spectrum examined in the farms. In the first analysis (1), all samples from all 116 farms were evaluated for the frequency of the bacterial pathogens. In the second analysis (2), the combined data for bacteria and viruses in 79 of the farms were examined for the simultaneous detection of more than one pathogen for neonatal diarrhoea.
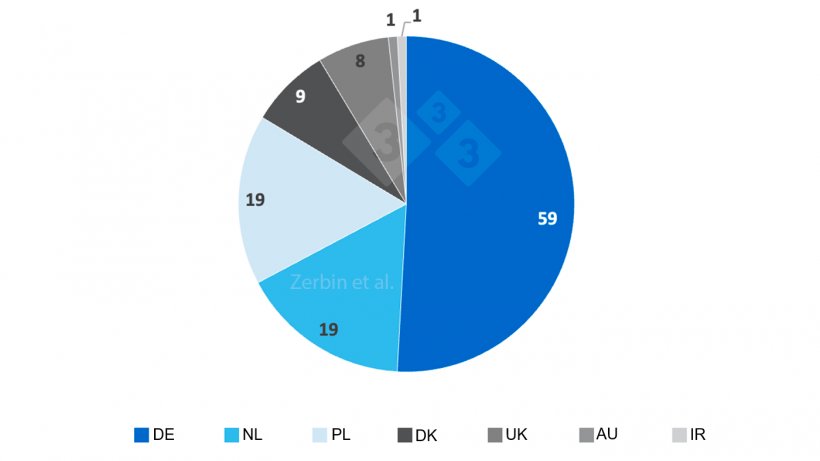
Most of the 116 farms (Fig. 1) were located in Germany (DE; n=59), followed by The Netherlands (NL; n=19) and Poland (PL; n=19), Denmark (DK; n=9), United Kingdom (UK; n=8), Austria (AU, n=1) and Ireland (IR; n=1).
The E. coli and C. perfringens isolates in both analyses were further typed molecular biologically in order to determine the occurrence of virulence associated genes and therefore evaluate their virulence potential (Strutzberg-Minder and Dohmann 2015). Additionally, the semi-quantitative bacterial content was analysed for E. coli, C. perfringens and C. difficile in analysis 1, based on their growth in culture (Tab. 1).
Results
Analysis 1:
In the bacteriological examination of the samples from all 116 farms, a total of 710 isolates were found (s. Fig. 2). Among them, the most frequently isolated bacteria were E. coli (48.6%), followed by C. perfringens (33.9%) and C. difficile (15.9%). The analysis of their semi-quantitative detection (Tab. 1) revealed that 99.4% of the E. coli isolates occurred in a moderate or high content, and even 96.7% of C. perfringens isolates. However, C. difficile was found only in a moderate (46.9%) and minor content (53.1%). Only 16% of the E. coli isolates showed haemolysis (Tab. 1) and even among the defined EC pathotypes and potentially virulent EC (Fig. 3), only 16% of the isolates were haemolysing, confirming that the property of haemolysis is not a sole characteristic of virulence.
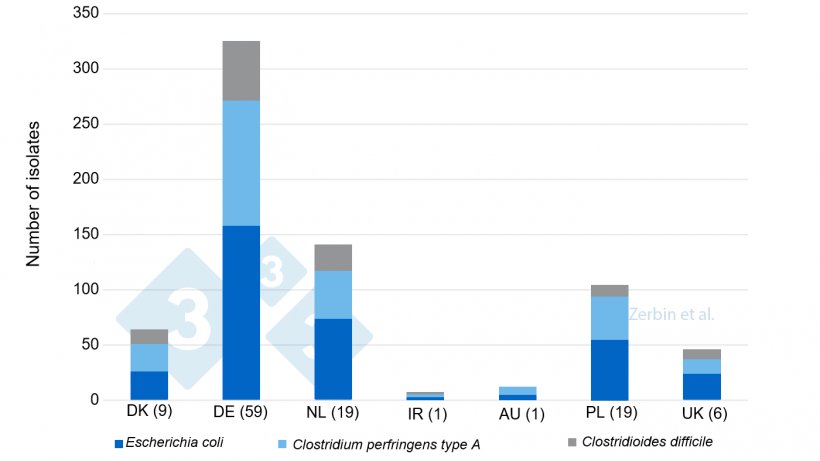
Table 1: Number (n) of culturally detected bacterial isolates in analysis 1, and proportions of semi-quantitative levels within each bacteria species.
| Pathogen | n | % | ||
|---|---|---|---|---|
| high content | moderate content | minor content | ||
| Escherichia coli | 345 | 77.1 | 22.3 | 0.6 |
| among them haemolysing | 57 (16.5%) | |||
| Clostridium perfringens | 241 | 46.9 | 49.8 | 3.3 |
| Clostridioides difficile | 113 | 0.0 | 46.9 | 53.1 |
After typing the E. coli and C. perfringens isolates and matching them to the single farms, a total of 276 E. coli and 117 C. perfringens isolates remained. They differed from one another with regard to their virulence-associated factors resp. toxin gene patterns within a single farm. Based on their virulence gene pattern, 19.9% (n=55) of these E. coli isolates could be assigned to one of the known pathotypes related with neonatal diarrhoea (EDEC, EPEC, ETEC, NTEC). Another 104 isolates (37.7%) carried various genes for fimbriae and adhesins as well as for toxins, and therefore can be classified as potentially virulent. Therefore, a total of 57.6% of the different E. coli isolates (n=159) were classified as virulent or potentially virulent for neonatal diarrhoea. Fig. 3 shows the distribution of the typing results for E. coli by country of origin.

The typing of the C. perfringens isolates revealed that they all belonged to C. perfringens type A (CPA). 90.6% of them (n=106) carried the coding genes for both the α- and ß2-toxin, and therefore showed a high virulence potential. The rest of the isolates (9.4%, n=11) only carried the α-toxin gene (Fig. 4).

Analysis 2:
Regarding the subgroup of 79 farms, the pathogens found most frequently in the farms were E. coli (96.2%), C. perfringens (87.3%), Rotavirus (74.7%) and C. difficile (46.8%). PEDV was detected in two farms only, TGEV in none. Therefore, the group of Coronaviruses was not considered in the further analysis.
Subsequent typing revealed virulent or potentially virulent E. coli isolates in 78.9% (n=60) of the farms, and CPA with ß2-toxin gene in 81.0% (n=64).
In 25.3% (n=20) of the farms, all four different pathogens could be found simultaneously, three different pathogens in 39.2% (n=31), two pathogens in 31.6% (n=25) and in 3.8% (n=3) of the farms only one pathogen was detected. Only in one farm none of the tested pathogens was found.
Among these the combination of E. coli with Rotavirus and CPA with β2 toxin occurred in most (n=19/ 24.1%) of these farms. The combination with E. coli, CPA and C. difficile occurred significantly less often (n=8).

E. coli as the most frequently found pathogen occurred mainly in combination with CPA with β2 toxin gene (64.6%) or rotaviruses (64.6%).
Discussion
The present study analysed the occurrence of relevant pathogens for neonatal diarrhoea in European farms, which voluntarily took part in a monitoring program for suckling piglet diarrhoea. The interpretation of the present results should be done carefully, as there is a certain selection bias in the sample design. The farms sent in the samples voluntarily, due to diarrhoea problems. Nevertheless, the data show realistic and clear trends concerning the pathogens causing neonatal diarrhoea that have been described in literature already.
Both analyses of this study confirm E. coli and C. perfringens as the main bacterial pathogens that occur in suckling piglet diarrhoea. The present data showed that almost 58% of the E. coli and almost 91% of the C. perfringens isolates can be considered as virulent or potentially virulent. In the second analysis, relevant E. coli and C. perfringens isolates could be found in approximately 80% of the farms each.
An interesting outcome is the very low evidence of Coronaviruses (PEDV and TGEV) in the samples of the study. Although they are mentioned as possible agents for neonatal diarrhoea, their proof in literature is usually low and diarrhoea caused by TGEV has become uncommon in Europe. The combined analysis of the different bacterial and viral pathogens in the 79 farms revealed simultaneous findings of two or more pathogens in almost 95% of the farms. This shows that neonatal diarrhoea in pigs rarely has a monocausal etiology. This should be considered with in terms of prophylactic and therapeutic measures.