Clinical signs of PRRS virus (PRRSV) infection can be diverse and therefore clinical diagnosis must be confirmed by laboratory examinations. Most often ELISA and RT-PCR are used for detection of antibodies specific to PRRSV or viral nucleic acid respectively.
Selection of a suitable diagnostic approach, depends on the diagnostic goal. Highly specific methods are preferable for monitoring of negative herds, while testing of semen from individual boars would require maximal sensitivity and methods detecting infection at earliest possible stage. In PRRS control program planning and execution, the best approach is to use combination of ELISA and PCR methods followed by DNA sequence analysis. Sampling protocol in a given farm has to be planned individually, depending of the diagnostic goal, and a farm organization. The simplest question, whether a farm is infected with PRRSV, can be answered by ELISA testing of 10-20 finishers. Detailed analysis of PRRSV circulation in a farrow to finish farm, or a multi-site production system, requires testing of large number of samples representing several age groups and all locations of a system, as they may represent independent viral ecosystems.

Interpretation of seroconversion or virus detection in farms where modified live vaccines are being used is challenging as (1) there are no marker vaccines on the market, (2) vaccine viruses can persist in vaccinated farms, and (3) at least for some time they can co-exist with wild type strains. In such farms DNA sequence analysis of amplicons obtained in different age groups of a farm is required for full understanding of PRRSV circulation.
Serological diagnosis based on ELISA is easy to perform and has generally good specificity and sensitivity when applied on a herd basis. However, sera of individual, especially adult pigs, sometimes give false-positive reactions. This constitute a problem in monitoring of negative populations and require re-testing of samples with different method (e.g. IPMA, IFA or another ELISA test of higher specificity), or resampling the pig population after 2–3 weeks. This aspect of ELISA performance is rarely addressed in evaluation of diagnostic competences. Low sensitivity of a given ELISA kit can be overcome by increasing the number of tested samples. Low specificity always requires application of additional tools.
Generally, any ELISA kit is useful for the detection of antibodies against either genotype of PRRSV, but kits employing antigens from only one genotype PRRSV are often less sensitive towards the second genotype. There are also kits available to discriminate seroconversion against the two genotypes but due to serological cross reaction between the genotypes the results should be interpreted with care. Discriminatory PCR is always recommended to confirm the diagnosis of farm co-infection with two genotypes.
PCR methods detect viral nucleic acids based on complementary binding of short synthetic oligonucleotides (primers) to a specific fragment of viral genome, which is later amplified in enzymatic reaction. In most widely used real-time PCR methods the product of a reaction is detected by monitoring of a signal emitted by the DNA probe after binding of specific sequence. Due to the nature of this reaction such signal may also be emitted in the absence of viral sequence, particularly in late cycles of PCR, which can resemble weak positive result.


Ellingson 2013, thesis, Iowa SU
However, the major concern regarding RT-PCR methods is related to high genetic diversity of the virus which may lead to accumulation of nucleotide mutations within genome fragments where primers and probes bind, and consequently to false negative results of PCR. Results of a comparative analysis of commercial RT-PCR kits performed by an OIE Reference Laboratory (Podgorska et al., 2015) indicated that some commercial RT-PCR kits failed to many PRRSV Type 1 strains originating mainly from Eastern Europe. Due to limited availability of sequences from Eastern Europe most of primers and probes in currently used methods were developed based on classic strains from Type 2 and Type 1 subtype 1, while Eastern European strains of subtypes 2-4 are genetically highly divergent (Stadejek et al. 2013). Moreover, some of them were proved to be more virulent compared to strains circulating in Western and Central Europe, so more attention should be given to ensure early detection in case of their spread.
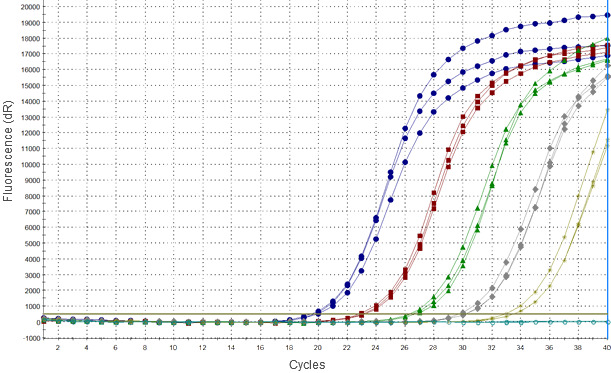
Currently, no single RT-PCR assay could be recommended as a universal method allowing for detection of all PRRSV strains with optimal sensitivity. Those observations highlight the necessity of a thorough assessment of currently used methods and their constant re-validation based on currently circulating strains. For this, a collection of representative PRRSV strains covering known genetic diversity of the virus has to be created, which is at the moment one of the most important aims of the reference laboratories for PRRS.








