E. coli (EC) – culture and serotyping
E. coli, a gram-negative, rod-shaped bacterium, occurs in a multitude of variants. In addition to commensal, naturally occurring strains found in the intestinal flora of all mammals, there are also pathogenic strains classified as intestinal (InPEC) or extra-intestinal E. coli (ExPEC). However, differentiation of these variants has long posed a diagnostic problem because different pathotypes or pathovars cannot be identified with simple phenotypical methods.

E. coli can be detected in swine relatively unproblematically by culture of suitable sample material (small intestine, feces) from animals with acute infection, on blood agar (Figure 1) or differential nutrient media, e.g., Gassner or MacConkey agar. In the past, typing of serotypes or serovars has successfully been used to differentiate E. coli isolates, due to the fact that certain serotypes are frequently associated with particular diseases (Table 1). It is therefore usual in veterinary medicine to determine the O and F antigens, by rapid plate agglutination using poly- or monovalent sera. (O antigens are components of the bacterial cell wall LPS and are also endotoxic; F [fimbrial] antigens are adhesins that facilitate host-specific adhesion to and colonization in the epithelial cells of the small intestine.) Although this method is quick, it is subject to error due to cross-reactions, masking of the target antigens, or the failure of fimbriae to develop in vitro, all of which can lead to false classifications. On the other hand, the O:K:H seroformula has for decades been and still is the standard for a complete determination of all serotypes, e.g. to determine not only the O antigens but also the K (capsular polysaccharide) and H (flagellar protein) antigens.
Table 1: Intestinal E. coli pathogens in swine
| Pathotypes | Diseases caused by E. coli | Frequently associated O antigen serotypes | Frequently associated fimbrial serotypes |
| Intestinal pathogenic E. coli (InPEC) | E. coli diarrhea in suckling pigs | O8, O108, O138, O139, O141, O147, O149 and O157 | F4, F5, F6 and F41 |
| E. coli diarrhea in weaner pigs | O8, O108, O138, O139, O141, O147, O149 and O157 | F4, F18ac | |
| edemea disease in weaner pigs | O138, O139 and O141 (O147, O157 and routinely not serotypeable) | F18ab |
E. coli (EC) – pathotypes and virotyping
Further developments in molecular biological methods and improvements in the molecular biological analysis of E. coli have led to the identification of many genes that code for virulence factors (Table 2): on the one hand, classic virulence factors with harmful effects (toxins), and those that directly or indirectly facilitate colonization and survival of E. coli in the host on the other. Identification of these genes, for example by PCR or array techniques, now makes it possible to determine the potential virulence of E. coli isolates and to distinguish between different pathotypes (Table 3). The identification of these virulence markers and thus their target genes is called virotyping.
Table 2: Examples of target genes detected in E. coli and the virulence factors (fimbriae, adhesins, toxins) for which they code
| Target genes | Virulence factors |
| faeG (F4) | F4 fimbriae |
| fanC (F5) | F5 fimbriae |
| fasA (F6) | F6 fimbriae |
| fedA (F18) | F18 fimbriae |
| fim41A (F41) | F18 fimbriae |
| fimH (F1) | type 1 fimbriae |
| fimA (F1) | type 1 fimbriae |
| papC | P fimbriae |
| aidA (AIDA) | AIDA-I autotransporter adhesin |
| paa | porcine adherence factor |
| eaeA (intimin) | intimin |
| eltB (LTI) | heat-sensitive enterotoxin |
| estA (STI) | heat-resistant enterotoxin |
| est B (STII) | heat-resistant enterotoxin |
| estA (EAST) | heat-resistant enterotoxin |
| stx2e | Shiga toxin variant 2e |
| cdtB | cytolethal distending toxin |
| cnf1 | zytotoxic necrotizing factor type 1 |
| iucD | aerobactin |
| escV | type III secretion system |
| pic | serin protease autotransporter |
Table 3: Target genes for the identification of E. coli pathotypes in swine
| Pathotypes | Fimbriae and adhesins (target genes) | Toxins (target genes) | Symptoms | |
Intestinal pathogenic E. coli (InPEC) |
enteropathogenic E. coli (EPEC) | fimA/fimH paa intimin |
escV pic cdtB |
diarrhea due to malabsorption |
| enterotoxic E. coli (ETEC) | F4, F5, F6, F18, F41 fimA/fimH AIDA paa |
STI, STII, EAST LTI Stx2e hemolysin |
diarrhea due to hypersecretion | |
| edema disease E. coli (EDEC) | F18 AIDA |
Stx2e hemolysin |
edema disease | |
| Shiga-toxin forming E. coli (STEC) | fimA, fimH intimin |
Stx2e EAST escV pic, cdtB hemolysin |
catarrhal, hemorrhagic diarrhea | |
| Extraintestinal pathogenic E. coli (ExPEC) |
septicemic E. coli (SEPEC) | papC | iucD | hemorrhagic septicemia |
| uropathogenic E. coli (UPEC) | papC fimA, fimH | cnf1, iucD hemolysin |
coliform mastitis in sows | |
However, pathogenic E. coli isolates always contain a combination of fimbriae and adhesins, in addition to toxins. Several toxins and adhesins can be present at once, and in different combinations. Determination of these various virulence markers and identification of their genes provides information about the pathotype of E. coli isolates from cell culture (Table 3). It must be stressed that hemolyzing E. coli are not the only ones that are pathogenic (Figure 1), and in fact non-hemolyzing E. coli in particular are frequently determined as the cause of clinical disease. Moreover, hemolyzing E. coli do not generally possess relevant fimbrial and toxin genes. Therefore all variants of the E. coli isolates identified by culture should be virotyped if possible in order to make certain that the responsible pathogen can also be identified. Information about the fimbrial types and toxins of pathogenic E. coli determined is also helpful in the selection of suitable vaccines with the corresponding antigens.
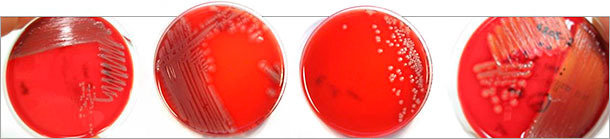
Figure 1: Different colony morphologies of pathogenic E. coli isolates on blood (Columbia) agar identified after virotyping by multiplex PCR. Not only hemolyzing E. coli (right) are pathogenic.

When interpreting diagnostic results it must be borne in mind that E. coli pathotypes can of course also be isolated from healthy hosts (latent infection), particularly from the gut habitat. Evaluation of diagnostic findings can therefore be made only in consideration of both the clinical findings for the animal in question as well as the concentration of the pathogens isolated with the identified pathotype.
Interpretation of virotyping becomes difficult when virulence markers are detected directly in the sample material, e.g. feces, without typing of individual isolates. Although this is technically possible and is provided on demand, when different E. coli pathotypes are present in the sample, one also obtains a mix of all detectable factors, potentially leading to the identification of false pathotypes and a combination of virulence markers which as such does not exist. Furthermore, it is also possible that virulence markers of other gut bacteria present in the feces sample will be detected, since these bacteria also have similar genes. In this case, apparent E. coli virulence markers will be detected which in fact are not virulence factors for the E. coli detected in the sample. Therefore particular care must be taken in the interpretation of the results of virotyping of samples.
In combination with the clinical and often necessary pathological examination (described in detail elsewhere) and with appropriate differential diagnoses, virotyping of E. coli isolates nevertheless brings marked improvement in the diagnostics of E. coli infections in swine.



