The diversity of influenza viruses is mainly explained by two closely interrelated aspects: On the one hand, the evolutionary mechanisms of these viruses –drift and reassortment (discussed in another article in this section)–; on the other hand, the epidemiological behaviour of the virus in each species, which has an important effect on the evolutionary mechanisms.
If we compare the evolution of these viruses in pigs with those present in humans in the perspective of genetic drift, two phenomena should be mainly highlighted; 1) given a particular strain, the virus varies antigenically faster in humans than in swine, despite equal rate of genetic mutation in both species, and 2) globally, the diversity of influenza viruses, is higher in pigs than in humans at genetic and antigenic level. Although these two facts may seem contradictory, they are perfectly compatible, as we shall see below.

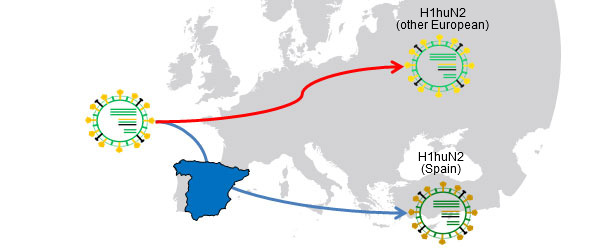
Figura 1. H1huN2 (1994) common ancestor; Geographical isolation → Drift causes variants country-related
Flu in pig herds may circulate as an enzootic disease as we have seen in previous articles. In other words, influenza virus can "reside" on farms. This situation is very common and involves the emergence of recurrent circulations (Loeffen et al., 2009, Simon-Grifé et al., 2012, Rose et al., 2013). In a paper published in 2001 (de Jong et al.), more than 20 avian-like H1N1 (H1avN1) isolates were compared genetically and antigenically. These viruses were isolated in the Netherlands from 6 swine influenza endemically infected farms. Based on the hemagglutinin gene (H), these viruses clustered in five groups that differed in their geno- and phenotype. It is important to note that each group corresponded to a farm (except one group including isolates from two farms). This work demonstrated that all isolates H1avN1 had a common ancestor, but endemic circulations on each farm forced them to diversify in five clusters by drift. In another study (Martin-Valls et al., 2013), held at CReSA between 2009 and 2012, 4 H1N2 virus were isolated. When analysed, it was observed that the H1 of all isolates belonged to the H1N2 type "human type" (H1huN2; most common European strain of swine flu), but clustering a distinct group within the H1huN2 cluster; so, it could be suggested that there had been an evolution and differentiation forced by geographical isolation in Spain. Later, we found that these viruses showed limited reaction with sera that reacted with high titres against a commercial H1huN2, evidence of antigenic drift (unpublished data). In short, genetic and antigenic drift occur in farms under endemic situations and by geographical isolation; this phenomena play an important role in the emergence of new antigenic variants. In humans, however, the situation is completely different. In this case we could say that, due to the overall immune pressure, viruses are antigenically forced to change in order to survive. Currently, the many and varied connections among countries around the world eliminate situations of viral isolation. Therefore, in humans there is an overall positive selection of those variants of influenza viruses able to escape the immune response; this phenomenon results in constant need of an update of flu vaccines in humans.
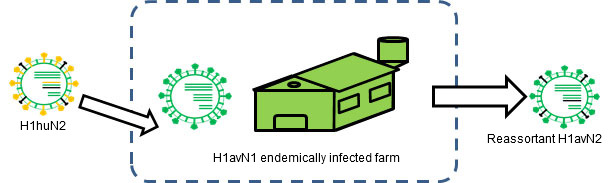
Figure 3. H1huN2 is transmitted into a H1avN1 endemically infected farm. → Reassortment occurs.
Reassortment can also be a source of antigenic changes. In humans, for example, reassortment has been one of the main causes of the last three pandemics. The pandemic viruses were reassortant that included at least a new antigenically different hemagglutinin. Since human population was naïve to these new reassortant viruses, transmission at pandemic level occurred. In pigs, as we saw in another article in this section, flu reassortments are frequent, and a big proportion of the swine influenza strains have been originated by means of this evolutionary mechanism. Examples are the H1avN2, an H1N2 cross-reacting with H1avN1 virus (Rose et al., 2013), or H1huN1, crossing antigenically with H1huN2 (Martín-Valls et al., 2013, Rose et al., 2013). For a reassortment to occur, two strains infecting the same animal at the same time are needed. Obviously, endemic situations are the best scenario for this to happen. In fact, when flu endemic situations have been evaluated, evidences of reassortment between different strains and even subtypes have been demonstrated (Martin-Valls et al, 2013; Rose et al., 2013).
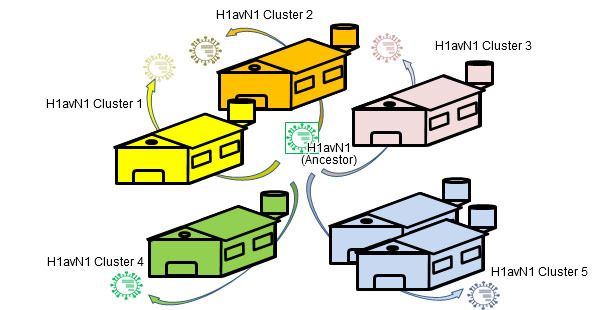
Figure 2. Endemic infections ease isolated evolution by drift on each farm → genetic and antigenic heterogenicity farm-related.
In conclusion, swine influenza viruses are diverse at genetic and antigenic level. This diversity is the result of the intrinsic evolutionary mechanisms of these viruses combined with epidemiological factors such as endemic circulation within a farm and/or geographic isolation. The interaction of all these factors has implications in the lack of immune recognition, resulting in a limited cross-reactivity in viruses sharing a common ancestor or belonging to a same subtype.



