The genomics revolution has been made possible by so-called ‘next-generation’ or ‘high-throughput’ sequencing technologies, which were outlined in the recent article ‘What do we mean by the genomics revolution?’ The principles of genomic selection are:
- DNA is analysed by means of a ‘gene chip’ to identify differences in the genetic code between animals called SNP’s (Single Nucleotide Polymorphisms).
- The SNP’s link specific segments of DNA to performance differences for specific traits in a specific population.


One of the greatest long-term potentials for genomic selection is to identify specific genes or genomic regions with a significant impact on pig health and to aid selection for disease resistance and/or disease tolerance. Several major research programs and collaborations are underway to investigate these areas and there is a growing database in the scientific literature.
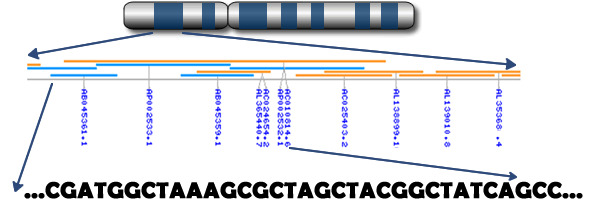
Figure 2. Genomics is identifying SNP’s for health traits (Source: Prof. C. Haley – Roslin Institute, Edinburgh)
For example, at the recent influential PAG (International Plant and Animal Genomics) meeting earlier this year, among key papers were the following:
- Researchers in the US have identified three markers that are significantly associated with lung lesions caused by pleuropneumonia and/or enzootic pneumonia. It is hoped that this will allow the identification of causative genetic variation involved with improved resistance to respiratory disease.
- Chinese scientists reported on the mechanisms of different disease status between the Landrace breed (susceptible) and a ‘local’ Chinese breed (the Tongcheng) which is resistant to PRRSV challenge. They observed different expression of the CD169 gene in the Tongcheng compared with Landrace after PRRSV infection. Some reported studies have shown that CD169 receptor improves antigen presentation to T cells.
- The PRRS Host Genetics Consortium (PHGC) was established by research groups in Canada and the USA to probe the role of genetics in resistance to PRRSV infection and related growth effects. Latest results from nursery pig studies identified a genomic region on porcine chromosome 4 (SSC4) with significant impact on variation in viral load and growth response following viral challenge. Further analyses and SSC4 region sequencing should identify markers that distinguish PRRSV resistant/maximal growth pigs from PRRS susceptible/reduced growth pigs.
- Further studies by the PHGC are studying transcriptome analysis of PRRSV-challenged pigs. For genes to produce proteins, DNA is ‘transcribed’ to messenger RNA (mRNA). The term transcriptome is used to cover all the mRNA that is used for protein production. This study will provide major insights to decipher genetic mechanisms for host response to viral infection by PRRS infection.
- Researchers at Iowa State reported on genome analyses that revealed many differentially expressed genes between pigs classified as persistent or low Salmonella shedders. They concluded that quantitative differences in IFN-γ levels explain the expression of most tested genes, and that the IFN-γ regulon is a source of genes whose expression levels two days post-infection can predict shedding outcomes. Such genes can now be further evaluated as candidates for development of predictive assays for shedding outcome in pigs.
- Chinese researchers studied immune responses in piglets for 18 haematological traits, seven lymphocyte traits and three cytokine traits using swine fever vaccine in two genotypes. They identified 12 significant SNPs which correlated with immune responses, suggesting a preliminary foundation for further identifying the causal mutations underlying overall immune response.
- Researchers from Nebraska reported on crossbred lines that were experimentally infected with PCV2b to identify major genetic variations that influence important indicators of disease progression and immune response to PCVAD. During a 28-day challenge period, weekly measures of weight gain, viral load and specific antibody levels were obtained. The initial results provide evidence of host variation in magnitude and time of immune response to PCV2b. Major clusters of SNPs that impacted viral load were located on multiple chromosomes including SSC6, SSC7 and SSC12. The region located on SSC7 also affected weight gain during challenge. The cluster of genes of the Swine Leukocyte Antigen class II is located in this region and could explain much of the observed phenotypic variation.

Figure 3. PCVAD – a key future target for genomic selection (Source: J. Mackinnon)
In addition to the above studies, work is underway to look at the genes/SNP’s involved in other major disease such as Foot and Mouth disease, African Swine Fever and Swine influenza. However, future selection for improved health will require the use of multiple markers and the availability of large DNA datasets from pedigreed populations with accurately measured health phenotypes that will identify associations between SNPs and health traits. The next article in the series, ‘When will genomics deliver?’, will look at this in more detail.