Detection of PRRS viral antigens
A definitive diagnosis of PRRS on sick pigs can be accomplished by detection of microscopic lesions characteristic of PRRSV in conjunction with demonstration of viral antigen in the tissues with lesions. The frozen tissue section fluorescent antibody (FA) test and immunohistochemistry (IHC) test can be used for detecting PRRSV antigen(s) in tissues (Figure 1). These tests are coupled with either monoclonal or polyclonal antibodies specific for PRRSV. The direct FA test on frozen tissue sections is inexpensive and rapid. The test is specific but is not always very sensitive. In particular, tissue quality (e.g., autolysis) affects test results. In contrast, IHC is useful for detecting viral antigen in formalin-fixed tissues. IHC is more sensitive than direct FA test but takes more time and is more expensive than the FA test.

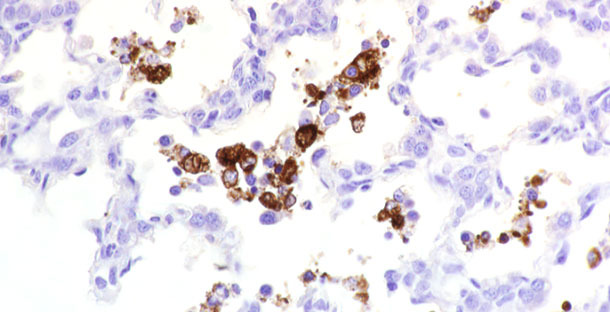
Figure 1. PRRSV antigen detected in tissue.
For direct FA examination, fresh or frozen tissues should be submitted. Tissues should be fixed in 10% neutral buffered formalin if submitted for IHC. However, prolonged fixation of tissues in formalin is detrimental to effective antigen retrieval for IHC. If a delay in performing the assay is expected, it is recommended that tissues be placed in alcohol after formalin fixation. Preferred tissues for these tests are heart, kidney, lung, lymph nodes, spleen, thymus, and tonsil. PRRSV antigens may also be detected in the adrenal gland, intestine, liver, and occasionally in the brain. When performing FA and IHC, monoclonal antibodies specific for antigens highly conserved among US and European isolates may be used, or monoclonal antibodies against less conserved epitopes may be used for evaluating antigenic differences between strains. If PRRS diagnosis is to be made by FA or IHC, due to the high specificity of monoclonal antibodies and the significant antigenic variability of PRRS virus, it may be best to use more than one monoclonal antibody in the assay to avoid a misdiagnosis.
Detection of PRRS viral genomic material
Polymerase chain reaction (PCR)-based tests have been developed for detecting PRRSV RNA (i.e., nucleic acid) in clinical specimens. Since the virus does not need to be isolated in cell culture to detect the viral RNA, PCR can provide significantly faster turnaround of test result than virus isolation. As a general principle, PCR-based assays are believed to be highly sensitive and highly specific.
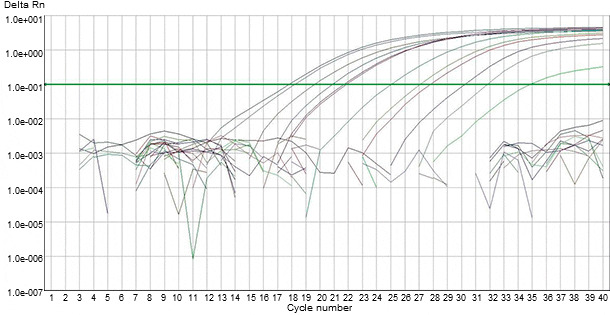
Figure 2. Detection of PRRS viral genomic material in clinical specimens by automated fluorogenic PCR-based tests.
Several types of PCR-based assays utilizing specific primers complementary to the sequences of different genomic fragment have been developed for detecting PRRS viral genetic materials in clinical specimens. With any PCR-based assay for PRRSV, the viral RNA needs to be extracted from clinical specimen and converted to complementary DNA (cDNA) using a reverse transcriptase enzyme (RT). Subsequently, the cDNA is exponentially amplified through the PCR process using a Taq polymerase enzyme and virus-specific primers. The precise match of the primers followed by exponential amplification is intended to produce both a highly sensitive and highly specific assay. More recently, automated fluorogenic PCR-based tests (i.e., RT-qPCR), have been developed for detecting PRRS viral genomic material in clinical specimens (Figure 2). These PCRs employ one-step amplification in one tube in which fluorescent probes bind to the PCR product as it is being made (i.e., “real-time” PCR). Fluorogenic PCRs are believed to improve the reliability and consistency of conventional RT-PCR for PRRSV detection (Figure 3).
Figure 3. Fluorogenic PCRs for PRRSV detection.
PCR-based tests have been used on a variety of clinical specimens, as well as oral fluid and environmental samples, for detection of PRRSV RNA. PCR is particularly useful for the detection of viral RNA in samples like semen or feces that are either cytotoxic for cell culture or cannot be evaluated by other methods. It has also been found useful for detection of PRRSV RNA in fetal tissues and thoracic fluids, i.e., samples in which PRRSV can be easily subjected to inactivation by the process of autolysis. The use of PCR assays has become more common both for the diagnosis of PRRS and to aid in herd monitoring/surveillance (i.e., screening of replacement animals, detection of carrier animals, and test and removal programs) or biosecurity (i.e., environmental testing, truck checking, semen testing).
PCR is expensive relative to other diagnostic methods. Furthermore, it should be kept in mind that a positive result on PCR indicates the presence of viral RNA and does not necessarily indicate the presence of infectious PRRSV. In addition, the performance of PCR testing among different laboratories may vary depending upon sample condition, sample processing, extraction technique, primers used, thermal cycling conditions, and the skills and experience of the technician performing the assay. Therefore, it is important for laboratories performing PCR to validate their assays, which should include estimates of sensitivity, specificity, comparisons to “gold standards”, proficiency testing results, and experimental or field studies of assay performance.








