Assays available:
It may be beneficial to review the great summary provided in Phil Gauger’s influenza article:

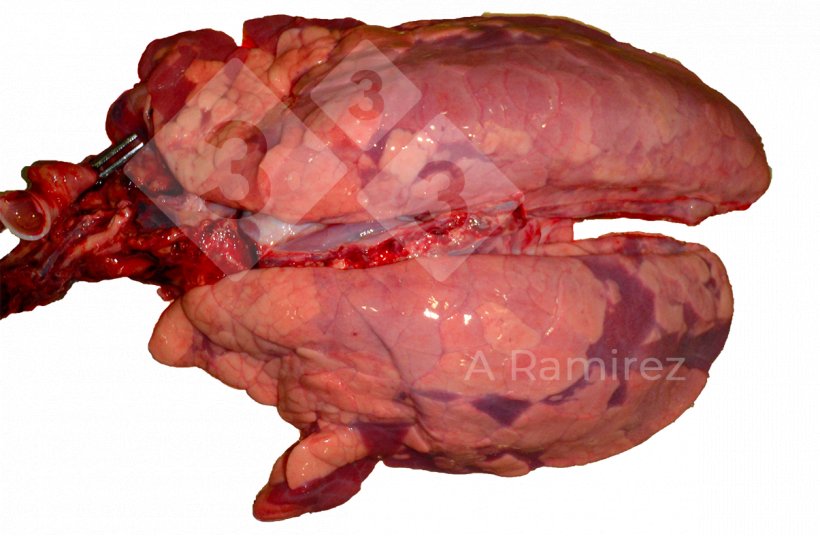
Gross pathology

- Evaluates the presence of tissue lesions which can suggest the presence of disease.
- Location of lesions:
- Cranioventral (especially apical and cardiac lobes) consolidation
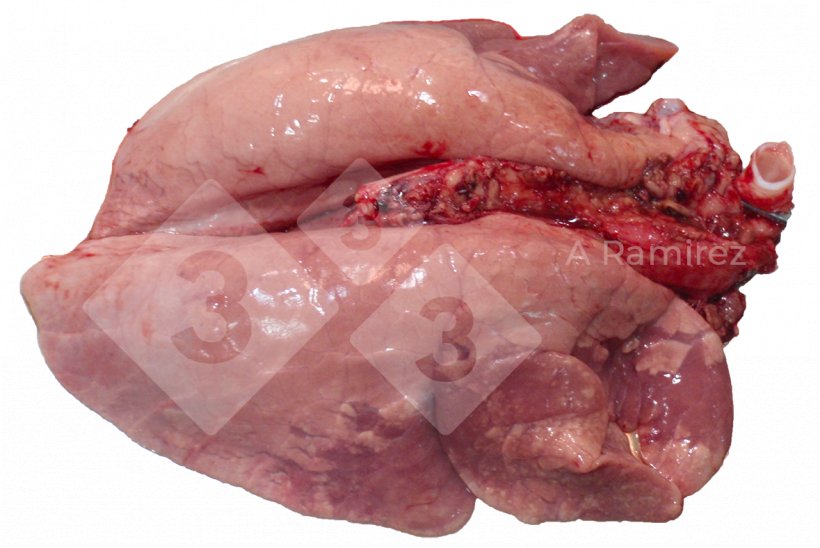
- Presentation can be diffuse covering multiple areas of the lung
- Some interlobular edema may be present
- Pros:
- Severity of lesions can relate to clinical significance
- Cons:
- Gross lesions not diagnostic (very similar to Mycoplasma hyopneumoniae)
- Often have secondary or co-infections
Histopathology
- Evaluates the presence of lesions which can confirm the presence of disease
- Sample types: Lung tissue
- Pros:
- Necrotizing bronchiolitis is usually considered pathognomonic for influenza A virus infection in pigs

Figure 3: Lung histopathology showing thinning of the bronchiolar epithelium, which is an example of necrotizing (photo source: Phil Gauger ISUVDL) - Characteristic lesions are present a few days after virus is eliminated from the pig especially when coinfections are present
- Necrotizing bronchiolitis is usually considered pathognomonic for influenza A virus infection in pigs
- Cons:
- Only evaluating a small tissues sample
- Uncomplicated lesions can resolve quickly (5-7 days)
Immunohistochemistry (IHC)
- Detects presence of viral antigen
- Sample types: Lung tissue
- Pros:
- Detects virus at site of lesion (good proof of causation)
- Target antigen is often a nucleoprotein which is conserved across influenza A viruses
- Cons:
- Only evaluating a small tissue sample
- Virus is only present for a short period of time (few days)
- Requires significantly more virus to be present than PCR
Virus isolation
- Isolates live virus
- Sample types: lung tissue, bronchial alveolar lavage, nasal swabs
- Pros:
- Traditional gold standard
- Isolate virus for use in vaccine development (autogenous vaccines) or serology testing (hemagglutination inhibition) to determine cross-protection
- Cons:
- Expensive
- Slow results
- Requires embryonated chicken eggs or Madin-Darby canine kidney (MDCK) cells
- Inoculation process is very labor intensive
- Often difficult to grow (many false negatives)
- Sampling handling from field to laboratory can impact virus survival
Polymerase chain reaction (PCR)
- Detects presence of specific sequence of viral nucleic acid (RNA)
- Sample types: nasal swabs, oral fluids, lung tissue, bronchial alveolar lavage
- Pros:
- Primers can be designed to target:
- Detection of all influenza A subtype viruses - target conserved nucleoprotein
- Detection of specific subtype virus groups: H1, H3, N1, or N2 subtype
- High sensitivity
- Early detection
- PCR quantification can be associated with presence of lesions
- Moderate cost
- Can often do pooling of swabs or tissue samples to lower cost while minimizing loss of sensitivity (especially regarding clinical relevance)
- Primers can be designed to target:
- Cons:
- High sensitivity – especially in oral fluids can detect presence of small amounts of virus in the herd making clinical interpretation of results challenging regarding the extent or severity of disease.
- Virus is only present in host for a few days (3- 5 days especially on nasal swabs)
- Cannot use blood or serum samples for PCR testing as virus stays in lung and is not systemic
Genetic sequencing
- Sequences virus’s genetic nucleic acid (RNA) segments
- Sample types: lung tissues, nasal swabs, bronchial alveolar lavage, oral fluids
- Pros:
- Can differentiate between subtypes into many different levels of clades
- H1: further divided into clades usually named using Greek alphabet (alpha, beta, gamma, etc.)
- H3: further divided into clusters usually named using Roman numerals (I, IV, etc.)
- Many of these relevant clades or clusters are described here.
- Can help differentiate new virus introduction from existing or past viruses
- Can help select vaccine strain based on subtype/clad
- Can differentiate between subtypes into many different levels of clades
- Cons:
- Expensive
- Usually only sequence HA gene
- Variability between subtypes/clades continues to grow and are becoming very complex
Enzyme-linked immunosorbent assay (ELISA)
- Detects presence of antibodies
- Sample types: serum
- Pros:
- ELISAS can be designed to target:
- Detection across subtype antibodies - target conserved nucleoprotein
- Detection of subtype specific antibodies – target specific hemagglutinin (referred to as H or HA) and/or neuraminidase (referred to as N or NA) proteins
- Animals remain positive for several weeks (start declining after 8-10 weeks)
- Can be used in chronic cases
- ELISAS can be designed to target:
- Cons:
- Specific antibodies detected and timing of detection vary slightly between the different commercial kits available
- Takes 10 to 14 days to become seropositive
- Unable to differentiate vaccine vs. wildtype infection
- Unable to identify specific subtype of virus
Hemagglutination Inhibition (HI)
- Detects presence of antibodies
- Sample types: serum
- Pros:
- Animals remain positive for several weeks
- Can be used in chronic cases
- Can be used to better determine proper time for vaccination by avoiding maternal antibody interference
- Can be used to determine cross protection between isolates
- Cons:
- Each assay is strain specific
- Requires isolation of specific strain to test for cross protection
- Takes 10 to 14 days to become seropositive
- Unable to differentiate vaccine vs. wildtype infection
Result interpretation
Gross pathology
- Positive: Confirmation of the presence of pneumonia
- Negative: Early cases may not manifest with extensive lung lesions
Histopathology
- Positive: Confirmation of disease
- Negative: Negative, or lesions could have been missed if testing wrong sample or too late after infection
IHC
- Positive: Virus is present at site of lesion
- Negative: Negative, or virus could have been missed if testing occurs too late after infection or the concentration of virus is too low (only present for a very short period of time)
Virus Isolation

- Positive: Confirmation of disease
- Negative: Negative, virus could have been missed if testing occurs too late after infection or simply unable to grow due to other contamination or poor sample handling (present but can be difficult to culture)
PCR
- Positive: Confirmation of disease
- Negative: Negative, virus could have been missed if testing occurs too late after infection or poor sample selection or handling
- Inconclusive: Too little virus present or have coinfection with more than one subtypes
Genetic Sequencing
- Allows for identification of virus subtype and clade
- Allows for matching vaccine isolate for better protection
ELISA
- Positive: Maternal antibodies or past exposure (> 2 weeks) to vaccine or wildtype virus
- Negative:
- Negative to vaccine or wildtype virus
- Infection too early to detect (10-14 days to seroconvert)
- Mismatch between assay and virus (subtype or clade)
HI
- Positive: Maternal antibodies or past exposure (> 2 weeks) to vaccine or wildtype virus
- Negative:
- Negative to vaccine or wildtype virus
- Infection too early to detect (10-14 days to seroconvert)
- Mismatch (subtype or clade) between virus used in assay and virus infecting pig
Scenarios
Growing pigs with sneezing and respiratory signs (acute or chronic):
- Collect 2-4 oral fluid samples from the group and test via PCR
- Collect 12 nasal swabs of infected pigs with clinical signs (have clear nasal discharge) and do 3 pools of 4 samples each testing via PCR
- Sequence one positive PCR sample to better determine cluster/clad
Determining timing of exposure and possible need for vaccination:
- Collect samples from 10-15 pigs at 3, 6, 9, and 12 weeks of age
- Two approaches to collecting:
- Cross-section – collect from different age groups at one time (quicker to obtain results).
- Longitudinal – collect from same pigs over time (results are more accurate).
- Serum samples test via ELISA.
- Collect 2-4 oral fluid samples from the group and test via PCR
- Sequence one positive PCR sample from each age group to better determine cluster/clad