Nucleic acid testing has become the standard for establishing the presence of pathogens in diagnostic samples. A common nucleic acid test procedure used in diagnostic and research laboratories is the polymerase chain reaction or PCR. PCR is a stepwise procedure that amplifies a minute fragment of DNA to the point that it becomes detectable either as a band on a gel strip or as a machine detectable fluorescent signal and it can be performed in a few hours. Significant expenditure in specialized equipment such as thermocyclers, a specialized laboratory, and trained and experienced personnel are required for good PCR work.
What is LAMP?
Loop-mediated isothermal amplification (LAMP) has emerged as a simple, rapid, sensitive and accurate method of DNA/RNA detection and amplification that requires no sophisticated equipment and is relatively inexpensive. The reaction is done in a single microcentrifuge tube. Required equipment for simple LAMP is a pipettor, disposable pipettes, and a dry bath heating block. A water bath or common incubator or even thermos bottles of hot water can suffice as a heat source but for ease of nucleic acid lab sanitation a dry block heater (Figure 1) is highly desirable.


In a typical application of LAMP, the test tube mixture will contain:
- Six DNA fragments called primers that match the DNA sequence to be detected.
- Specific polymerase enzyme (e.g., BST DNA polymerase)
- Buffers
- Supporting reagents
- An indicator dye (pH indicator, or direct DNA dye)
- Reverse transcriptase for RNA viruses
- The sample to be tested.
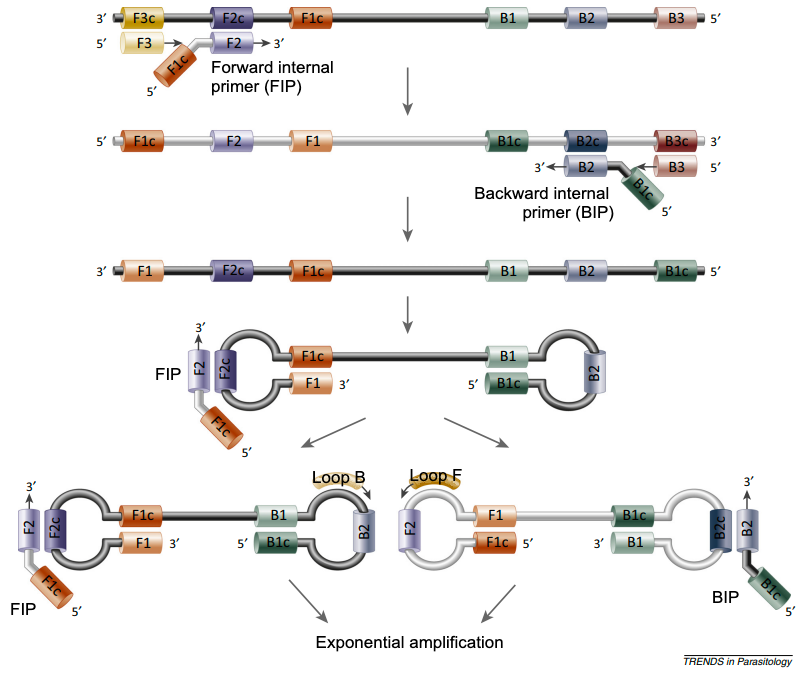
This mixture is incubated at 60-65°C for 30 minutes. If DNA matching the primers is present in the sample, a series of dumbbell shaped loops of DNA are constructed. The loops become more complex and more numerous as the amplification reaction proceeds (Figure 2).
The polymerase amplification produces magnesium pyrophosphate as a by-product that causes turbidity (cloudiness) in the tube. Indicator dyes that bind to DNA directly give a visible spectrum or UV fluorescent signal. One LAMP system uses phenol red pH indicator which changes from pinkish red to yellow.
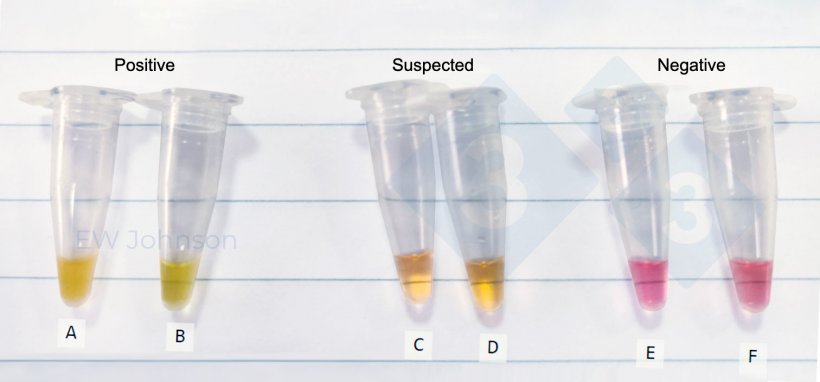
Figure 3 shows the result of a LAMP assay for African swine fever (ASF)- Tube A contained extract from the spleen of a sow that died from ASF. The high virus DNA content generates an opaque yellow colour. Tube B was an oral fluid swab with the typical lower ASF virus load from the same sow. Turbidity in the tubes A & B is evident by the obscuring of the background line. C to F are suspect and negative tubes.
There are some variations on the LAMP reaction as thematic widget. The test can be applied in sealed 96 well microplates with colourimetric or fluorescent dyes or measures of turbidity to give a quantitative and automated result. Filtered photocells in the heat block and a suitable LED could also automate the semi-quantitative real-time fluorescent reading of microcentrifuge tubes. Cell-phone camera spectrophotometry might resolve suspect tubes into binary logistic yes/no in some cases where human eyes can’t quite resolve the dilemma to our satisfaction.
The LAMP reaction can be done on-site in the field for practical “Point-of-Care” results with minimal waiting. Our lab constructed and verified inexpensive LAMP assays (from “scratch”, and from “kits”) for Classical Swine Fever, (CSF), Porcine Circovirus 2 (PCV2), and ASF (as above) as proof-of-concept (for CSF) that one could screen a batch of breeding animals before introduction quite cheaply for persistent infection by LAMP testing.
The advantages of LAMP are the rapid test results, minimal training required, low investment in equipment, small laboratory footprint, and low cost per sample overall. While costs may differ, the cost of LAMP is generally presented as about half the cost of PCR and it is further reduced for simple matrices where a nucleic acid extraction step is unnecessary. Labs that wish to invest in developing LAMP from scratch with the relatively cheap reagents and home-grown primer sets potentially avoid the built-in higher cost of a ready-to-use kit but face those costs associated with development, verification, and certification.
The sensitivity and specificity of LAMP is comparable to that of PCR and it can yield results in 30 minutes. Some jurisdictions have approved commercial LAMP test kits for ASF and human Covid-19.
Disadvantages of LAMP might include its unfamiliarity among some academic and research workers and some skepticism of the method. LAMP requires 6 (up to 8) primers for each gene to be detected and the generation of primers is more challenging than conventional PCR, but online tools are available. Multiple primer sets in the mix may be required for highly mutable RNA viruses like PRRS, but this has not precluded its utility. LAMP does generate a lot of DNA and the positive tubes generally should not be opened after the test to avoid lab contamination. LAMP amplicons are not readily amenable to useful sequencing and the lab may need to arrange conventional PCR or other methods for LAMP-positive samples when gene sequences are required.
One could easily envision LAMP application in livestock sale barns, animal collection points, and in slaughterhouses as well as in primitive and remote settings where one would not take sensitive and expensive equipment. Where sophisticated facilities or even electricity are unavailable and binary yes/no answers are imperative to human action in real time on the spot, LAMP could provide a clear way forward. The low cost and lack of need for equipment has hindered the proliferation of LAMP somewhat, since companies and university labs may prefer expensive physical facilities, and the marginal returns on investment may be more important than actual cost for fee-based health care services.
As the world moves toward control and elimination of important endemic transboundary diseases such as African Swine Fever (ASF), PRRS, and Classical Swine Fever (CSF), the availability of a rapid, convenient, cost-effective test has potential utility.
“Perhaps to be too practical is madness.” - Miguel de Cervantes Saavedra (1547-1616), Don Quixote.