In our previous discussion we highlighted how challenging it is to conduct scientifically sound studies on the swine microbiota. Despite this we have seen an increasingly larger number of studies published in this topic (Figure 1). As more information becomes available, how can you make sense out of these data – and apply it at the barn?

Step 1 – Know your terms.
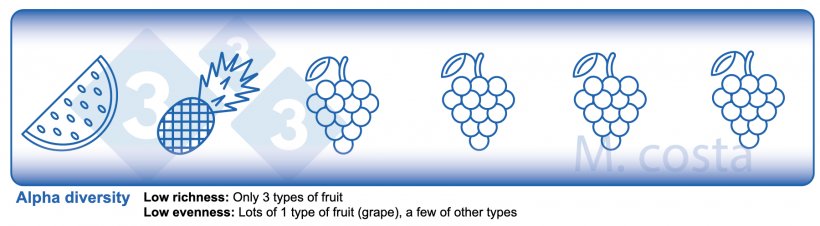
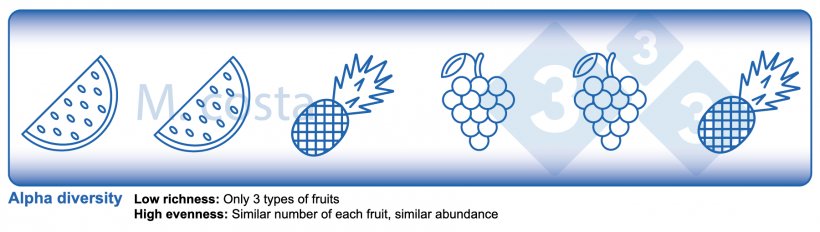
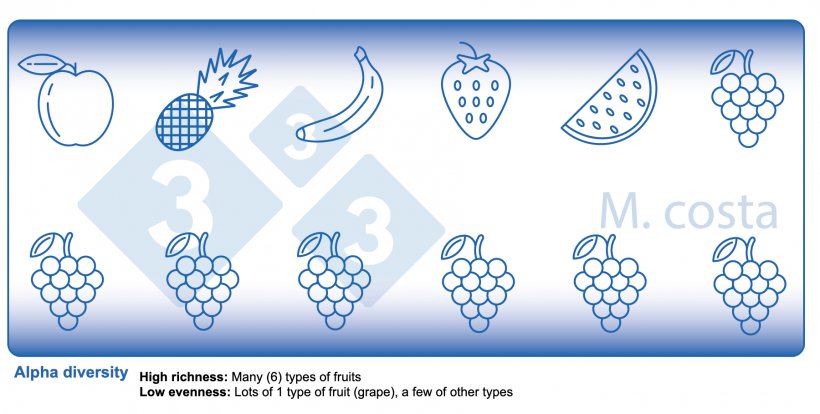
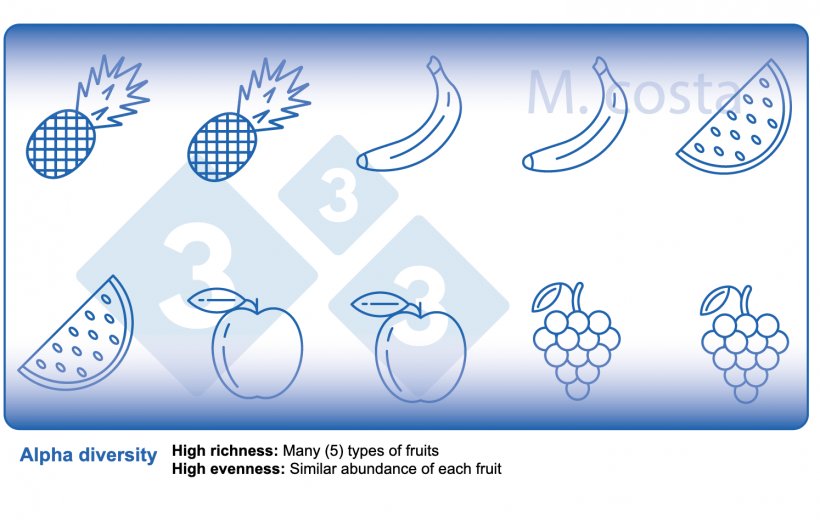
There are some key metrics used in microbiome studies that facilitate our understanding of the results. Many are borrowed from “macro” ecology – after all, a community of bacteria in the gut is not much different from a school of fish in the sea. They both have relationships with their environment and other beings that share their space/habitat. Therefore, understanding these metrics will allow proper interpretation of microbiome-focused experiments. In general, such studies focus on clarifying what microbes are present/absent in a given sample – they answer the general question of “who is there” and “how many of you are there”? The different species/types of microbes in a given habitat (e.g. the gut) are often referred to as the community diversity. This diversity of entities (e.g. fruits, or bacteria) in a single habitat (e.g. feces from nursery pig #495 on day 3 post-weaning) is called alpha diversity (Figure 2) and takes into account two main aspects:

- Species richness: how many different types of bacteria/species are there.
- Species evenness: the distribution (abundance) of these species, or is one organism predominant (more abundant) over others.
There are many indices used to measure alpha diversity, the most common being the Shannon, Chao1 or Simpsom’s index. Each one of them reflects different aspects of alpha diversity. You will see these indices often in microbiome studies, reflecting the within-sample diversity.
When comparing the bacterial community of two habitats (e.g. fecal samples from pigs treated vs not treated with antibiotics), we investigate their beta diversity - the similarity between samples. Do these communities have many things in common (e.g. can we find the same bacteria in fecal sample A and B, Figure 3)? Are they similar or dissimilar? Again, there are many indices used to measure this, such as Unifrac, Jaccard, Bray-curtis dissimilarity, etc. These take into account different aspects of community diversity, such as the (phylo)genetic relationship between the microbes, or their abundances in each sample.
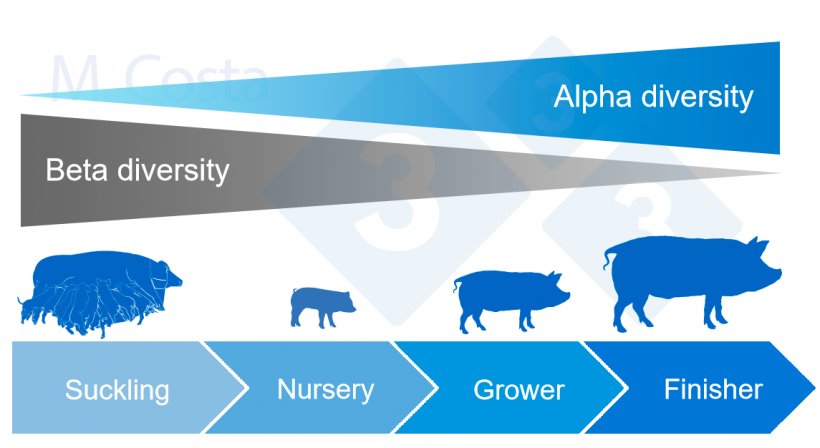
In general, gut microbial alpha diversity tends to increase, and beta diversity tends to decrease over the lifetime of a pig (Figure 4).
Step 2 – Did the study include the appropriate controls?
When it comes to microbiome studies, all efforts should be made to minimize the effect of contaminants. Microbial DNA is literally everywhere (even if the microbes are dead, their DNA is there) and the current DNA sequencing technologies can detect very little amounts of it (as discussed in Part 1). Therefore, studies not only must include the appropriate experimental groups (e.g. pigs either treated or not treated with antibiotics in the same batch, eating the same diet, etc…), but also the appropriate technical controls (empty collection tubes, sequencing kits without samples) to help identify contaminants and remove them from the analyses. If these are not disclosed in the study, any conclusions could be flawed.
Step 3 – Biological significance and beyond the P-value (or: did it really affect the animals?)
It is somewhat easy to disturb a microbial community structure. The gut microbiota, for example, is susceptible to changes in diet, physical space, and antibiotics. Interventions can easily cause significant changes to the community composition (both alpha and beta diversity). Very important work was conducted in the past decades to confirm that. However, evidence of biological significance is key. A lot of the “work” microbes do is redundant. For example, many members of the gut microbiota can produce the same short-chain fatty acids. Therefore, changes in the microbial community composition (or “who is there”) is likely less important than what they are doing. It is important to look for indirect evidence that the latter was affected by the intervention, and this may come in different ways: growth rate, resistance to disease, intestinal barrier function. It really depends on the study, but it should be reported.
As with any other cutting-edge technology, some important aspects may be lost when translating research from the lab bench to the farm. As microbiome modulation and its applications in swine production become clearer, we should see new strategies to help boost performance and disease resilience.





