There is a growing number of breeding herds endemically infected with emerging wild-type variants of PRRS virus in many countries. At the early stages of PRRSV outbreaks, there is production impact all the way from the breeding herd to finishers. However, as herd immunity establishes, the infection becomes minimum to subclinical in the breeding herd. The clinical signs at that stage are more evident in the growing pig herd, especially toward the end of nursery when the maternal immunity wanes.
Close monitoring of PRRSV infection is crucial to ensure the herd is making a timely and complete recovery of infection and clinical signs. In response to this need, there have been many new developments in monitoring & surveillance systems, including diagnostic and clinical approaches.

Diagnostic monitoring
According to the SDRS data, there has been a fundamental shift within the last decade from using serum samples to population-based samples for PRRSV RNA testing. Oral fluids are the primary sample type utilized in grow-finish pigs to screen for viral activity. In breeding herds, processing fluids (PF) are the most used sample, followed by oral fluids and tissues.
Processing fluids (PFs) are a practical sample to screen PRRSV activity in breeding herds doing physical castration of piglets. It covers 2-7 days old suckling pigs, with many farms submitting one pooled sample from piglets processed during the week. It is a common approach to collect PFs from as many litters and rooms as possible, keeping samples refrigerated or frozen at the farm, and submitting daily samples for one pooled PCR test to represent that particular week. Once the results start to test PCR-negative, the veterinarians may open the pools testing the individual days separately for increased sensitivity.
Once processing fluids start testing consistently negative by PCR (i.e., 4-6 weeks in a row), the main question is whether the weaning age population remains negative. In that situation, if the virus is still circulating in the herd, the prevalence will likely be very low (≤ 3% prevalence). Thus, a highly sensitive monitoring scheme is needed. That is when family oral fluids (FOF) come into place. FOF consists of hanging a rope where both the sow and the respective litter have access to it, leaving oral fluid samples representing ‘the family’. Sample size calculators have been reported to guide people in establishing the number of FOF samples and the intensity of pooling these depending on the farrowing room size and desired confidence level.
Another recently reported approach is using tongue-tip fluids from dead pigs. This method is based on taking samples from dead animals, where the virus is more likely to be there, and is very efficient compared to live animal sampling methods.This risk-based sampling method is handy when there is an unexpected drop in Ct values of PCR results from PF, serum, or FOF samples. Also, when there is an atypical long duration of PCR-positive results (i.e., 40 or more weeks after the PRRSV outbreak). Sampling the tongue tips from stillborn piglets allows understanding if the infection is coming from the breeding herd (i.e., vertical transmission) versus the farrowing house (i.e., horizontal transmission). Dr. Isadora Machado et al. have recently reported that the probability of PRRSV RNA detection in tongue tips from stillborn pigs was similar to or higher than that of processing fluids, serum, and family oral fluids.
Moreover, a recent promising sample called tonsil-oral scraping (TOSc) has been reported by Dr. “Jack” Li Peng. In summary, TOSc consists of using an insemination rod to sample the tonsillar area of sows, collecting a viscous sample. His preliminary data comparing PCR results from serum, tonsil scraping, and TOSc with 30 sows showed higher positivity in TOSc compared to other sample types. Studies are being performed at the time of this writing, and results should be available soon. Another advantage of the TOSc sampling is that it does not require straining sows, making the sampling process to take place at a relatively high speed when compared to the conventional tonsil scraping method.
Table 1: Monitoring techniques on population based samples.
| Sows and gilts | |
|---|---|
| Sample | Oral fluids |
| Objective/Why? | Understand PRRSV shedding |
| When? |
|
| At farrowing | |
| Sample | Tongue-tip fluids from stillborn pig |
| Objective/Why? |
|
| When? |
|
| At processing | |
| Sample | Processing fluids |
| Objective/Why? | To screen whole-herd viral activity in farms performing physical castration |
| When? | At any given point in time; PF are great to screen the herd for PRRSV circulation. When physical castration is not performed this can be substituted to tongue tips from pigs who died in the first week of life. |
| End of lactation (weaners) | |
| Sample |
|
| Objective/Why? |
|
| When? | FOF is implemented before weaning; OF within a few days post-weaning. OF is an alternative to FOF in pig flows from single sow farm sources operated in all in-all out flow. PCR-negative results then imply that the sow farm is weaning PRRSV-negative piglets. |
| Finishers | |
| Sample | Oral fluids |
| Objective/Why? | Assess viral activity |
| When? | Ideally, every 4-5 weeks to assess viral shedding. PCR-positive may be followed-up by other tests (e.g., sequencing, or vaccine-specific PCRs) to determine wild-type versus vaccine virus. |
Clinical monitoring
The diagnostic monitoring is very accurate with excellent specificity and sensitivity. However, it is usually not performed on a daily manner in most herds. To fill this gap, and keep the breeding herd on watch 24/7, producers are taking advantage of clinical monitoring using available data streams on the cloud. Algorithms based on statistical process control (SPC) are available to establish ongoing automated monitoring of selected parameters using farm-specific baseline. Examples include the number of aborts, number of sows off-feed in the gestation phase, and neonatal losses (stillbirths and mummified fetuses). When there is a significant increase in one or many of these parameters, alerts are issued to pre-registered farm users, who can run ad hoc diagnostic tests to determine the cause of variation.
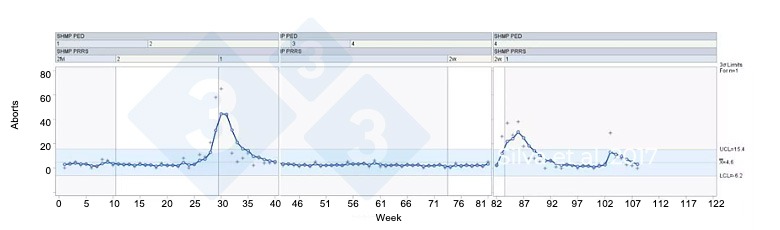
Figure 1. Applied SPC to detect herd level signs of PRRS. Silva et al. 2017.
Final considerations
Programs such as SDRS provide great information at the macro-epidemiological level. Such information is dynamic and evolves quickly over time and space. Population-based sampling in pig populations, including oral fluids, processing fluids, and tongue tips, allows for characterizing the PRRSV activity in the room (or barn) level in a practical fashion. The TOSc samples, if validated, represent a significant step to allow screening gestating sows for PRRSV. Each sampling approach serves to address a specific question. For example:
- processing fluids are great for screening whole-herd viral activity
- family oral fluids are to verify the PRRSV activity in the weaning-age pigs
- tongue tips of stillborn piglets are for assessing vertical transmission.
In addition to the diagnostic monitoring tools, producers should take advantage of available clinical data to keep the herd monitored 24/7, and thus being able to early detect disease outbreaks, allowing rapid response and adjust bio-containment measures as needed to prevent further pathogen spread to other swine populations having epidemiological connections with the herd. Altogether those sampling approaches equip the producers and respective veterinarians with several tools to understand where PRRSV stands in each herd.





