Historically, pigs have been considered as a “mixing vessel” with potential to adapt influenza viruses from other animal species (eg birds) to humans. This fact, in part, is due to the presence of two types of cellular receptors in the respiratory tract of swine, the α-2,6 and α-2,3. While the first type of receptor (α-2,6) allows binding and infection by human influenza viruses, the second (α-2,3) allows binding and infection by avian influenza viruses. Nowadays we know that humans can be directly infected by influenza strains of avian origin since they also have α-2,3 receptors. Still, pigs are a source of generating new strains with zoonotic potential. A clear example is the H1N1 virus that caused the 2009 pandemic (H1N1pdm). This virus was generated as a consequence of several genetic reassortment events including avian, human and swine influenza viruses. These reassortments probably occurred in the US swine herds the previous 10-11 years. At least four influenza viruses played a role in the generation of the H1N1pdm; one avian virus belonging to the American lineage, one human virus (H3N2 seasonal) and, finally, two different swine viruses, one American (H1N1 classical swine, descendant of the virus responsible for the Spanish flu of 1918) and one European (H1N1 "avian-like" swine).

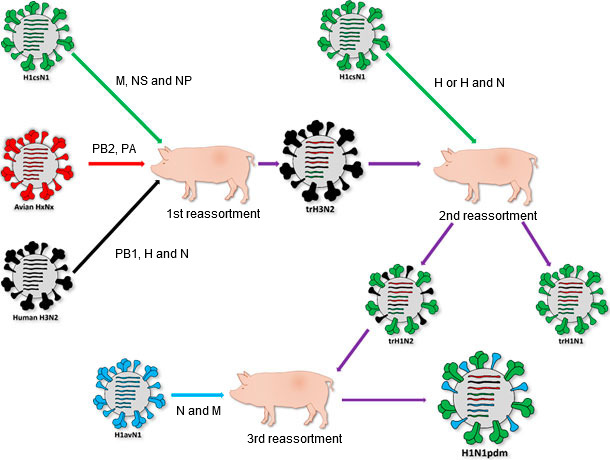
Origin of H1N1pdm. As it can be seen in the figure, de H1N1pdm strain was originated from at least three reassortment events on different moments. The first of them occurs between 1998 and 1998. This reassortment event generates the triple reassortant H3N2 (trH3N2). Three viruses were implicated on this event, a swine H1csN1 (H1N1 classical swine), an avian virus belonging to the American lineage and, finally, a seasonal human H3N2. The different given genes from each parental strain are indicated in the rows. The second event occurs son after. In between of 1999 and 2000, new reassortment between the H1csN1 and the trH3N2 generates two viruses, the triple reassortant H1N1 (trH1N1) and the triple reassortant H1N2 (trH1N2). Finally, near to 2009, the last reassortment is produced between an Eurasian “avian-like” swine H1N1 (avH1N1) and the trH1N2, generating the H1N1pdm and starting the pandemic.
Shortly after the start of the H1N1pdm pandemic, the virus was detected in a pig farm in Canada. This event is considered as the first reintroduction of the H1N1pdm in pigs. This reintroduction was probably produced by an infected worker. The impact of the respiratory outbreak in this herd was what would be expected from an outbreak of flu in pigs (low mortality and self-limitant). However, even though the animals did not pose any risk to human consumption, no slaughterhouse wanted to carry out the kills of these animals. Consequently, the farmer had to sacrifice the animals on the farm, assuming all costs. During the course of the following years, the H1N1pdm virus was detected in pigs of countries such as China, Germany, France, Italy, Spain and USA among others. Special cases are Norway and Australia. Both countries, which until 2009 were free of swine flu, suffered several outbreaks caused by the strain H1N1pdm in pig farms. In addition, the virus H1N1pdm has been a source of genetic reassortments with other swine influenza viruses worldwide, which has generated new strains.
How often genetic reassortments of influenza viruses occur in pigs? It appears that they are relatively frequent. In fact, a study published in 2012 (Lycett et al., 2012, J Gen Virol, 93:2326-36) estimated that reassortment events of swine influenza viruses occur every 2-3 years in viruses from Eurasian origin. In another study carried out in Spain (Martín-Valls et al., 2013, Vet Microbiol 170:266-77), 11 of 14 swine influenza strains with the genome fully characterized showed evidence of genetic reassortments. Even though in most cases reassortment occurred among swine strains, one isolate was product of the reassortment between human and swine influenza strains.
An interesting fact is that most of European and North American swine influenza strains contain part or whole genomes of human influenza viruses. While it is true that the swine-to-human transmission has been described in several countries, and in some cases, like that of the 2009 pandemic, had a great impact, human-to-pig transmission is much more common. This apparent increased susceptibility of pigs to infection with human influenza strains is due to one simple reason; production flows in swine can continuously generate pigs susceptible to infection (young animals, new replenishment, etc.). So, if a person infected with a human influenza virus enters into a swine farm, the probability of infecting the animals is very high. Although these animals develop immunity against this human influenza strain, after relatively short time new susceptible animals will be available. Therefore, the farm as a group would not be fully protected against infection with this same strain of human influenza. Conversely, if a worker of a pig farm becomes infected by a swine influenza strain, he would generate immune-protection that would prevent him from getting re-infected by the same strain in the future.
In conclusion, pigs can act as a source of new influenza strains with zoonotic potential, but the increase of the genetic diversity of the swine influenza viruses is largely due to the introduction of influenza strains of human origin. Therefore, it is highly recommended that all staff that has frequent contact with pigs (veterinarians, farm operators, abattoir workers, etc.) should be vaccinated against influenza and, of course, if potentially infected by human influenza virus, contact with animals should be avoided.



