There are two most widely used diagnostic methods to detect PRRS, ELISA and PCR. In this article we will focus on PCR, the method that can detect the virus in virtually any material. It relies on a cyclic DNA polymerisation of a DNA, or as in case of PRRSV a cDNA fragments flanked by short sequences to which specific DNA primers bind in each PCR cycle. The sensitivity and specificity of a diagnostic assay employing PCR depends on the fitness of these primers to their target and the thermal cycles employed for cyclic amplification. Ideally, they should be complementary only to the genome of a given virus species, and not able to bind to any other microbe or host DNA sequences, still, being able to detect genomes of all strains, or variants of that virus species. So, they target the most conserved fragments of the genome.
PRRSV is one of the most challenging pathogens to develop highly sensitive and specific PCR assays. Currently known PRRS viruses belong to two species, PRRSV-1 and PRRSV-2. Both PRRSV species are spread globally but PRRSV-1 dominates in Europe while PRRSV-2 dominates in America and Asia. It can be assumed that the genetic diversity of a dominating PRRSV species is higher than the one having lower prevalence. In effect, PCR assays being used for example in Europe or America, may be better validated, and better suited, to detect PRRSV-1 or PRRSV-2, respectively. As a consequence, some strains of the “minor” species in a given area, or a continent, could remain undetected, and their role in PRRS pathogenesis, underestimated. This is particularly important if in farms where PRRSV-1 and PRRSV-2 co-circulate. Great genetic diversity of PRRS viruses requires constant update of sequences of PCR primers and probes to follow the continuous evolution of the virus and discoveries of completely new genetic variants. This is especially true for PRRSV-1 which extreme genetic diversity and low availability of reference viruses from Russia, Belarus and Ukraine poses a threat of compromised sensitivity of commercial PCR kits towards the East European genetic subtypes of PRRSV-1.

Currently real time PCR variant is mostly used in routine diagnosis. There are several real time PCR methods that differ in their principles but the most widely used is a method with so called TaqMan probes. A TaqMan probe is a short DNA fragment (oligonucleotide) labelled with fluorophores, which binds the DNA target between the flanking primers. If binding occurs the probe is lysed during DNA polymerisation and fluorescence is recorded by the PCR instrument. Real time PCR is particularly well suited for multiplexing of the diagnostic targets. It means that in a single PCR reaction (or a single test) 2-3 different pathogens can be detected through the emission of fluorescence of 2-3 different dyes. So, simultaneously both PRRSV species can easily be detected, and differentiated in a samples from a single animal, pen, or a farm.
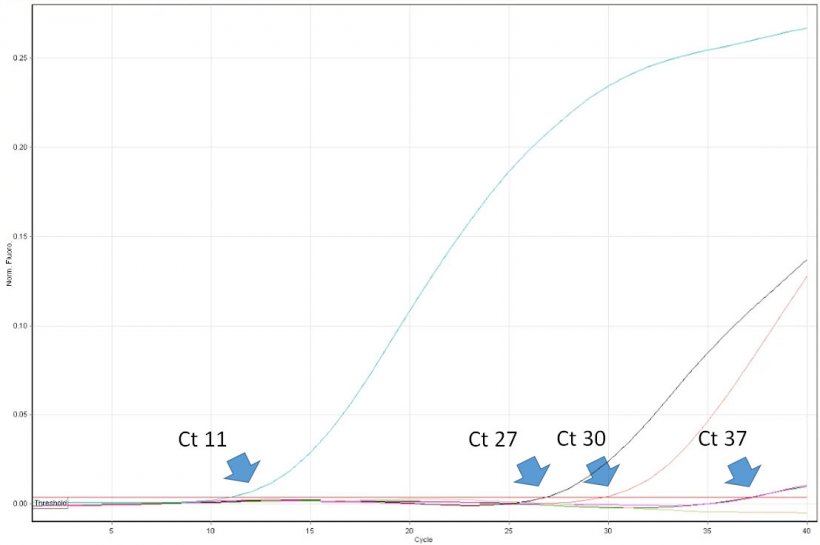
The interpretation of PCR results is deceptively easy. Veterinary practitioners have to be aware of some limitations of the method and be able to critically interpret its results. For example, positive result of PCR proves the presence of a target nucleic acid (e.g. of PRRSV) in the sample. However, it does not prove that the sample contained infectious virus. Real time PCR allows for virus quantification in the sample. Its results are expressed as a “threshold cycle” (Ct). It is the first cycle number in which the PCR instrument detected fluorescence emission from the reaction tube. The lower the cycle number, the higher the copy number of the target DNA (read as a number of virus copies). For example a sample with Ct 20 can be considered highly positive, and a sample with Ct 35 low positive. However, in some cases fluorescence is detected only in very late cycles of real time PCR, e.g. >38. How such high values can be interpreted? Unfortunately not always such result indicates the presence of a very low number of virus copies in the sample. The, so called, “late-risers” may be a result of a spontaneous break up of a probe after many thermal cycles of the PCR, which mimic enzymatic nucleolysis caused by DNA polymerase. So, it can occur also in the absence of the virus nucleic acid in the sample. The importance of the correct interpretation of “late-risers” is highest while monitoring PRRSV free populations. The follow up diagnosis, including ELISA, must be performed. Unfortunately, some diagnostic laboratories in Europe do not provide the customers precise Ct values of real time PCR. Instead the results are expressed only as negative or positive. Usefulness of such reports for practitioners is limited and in some cases may lead to wrong decisions regarding PRRSV control protocols.
Practical considerations
- None of the current PCR methods is able to detect all PRRS viruses.
- Negative result of PCR of a few random samples from a farm does not prove the freedom from PRRSV infection.
- Positive result of PCR proves the presence of PRRSV nucleic acid in a sample but not necessarily its infectivity.








