Swine influenza is a highly contagious respiratory viral infection of pigs that has become enzootic in areas densely populated. It can have a significant economic impact in an affected herd and may pose a threat to the human population, as swine influenza viruses (SIV) are zoonotic pathogens. Ongoing knowledge of SIV strains is indispensable for the prevention and control of the disease in pigs and to provide a baseline for risk assessment of interspecies transmission, in consistence with pandemic preparedness.
SIVs belong to Influenzavirus A genus, Orthomyxoviridae family. Their genome consists of eight unique segments of single-stranded RNA of negative polarity. They are subtyped according to their major antigenic determinants, the surface glycoproteins hemagglutinin (HA) and neuraminidase (NA). Emergence of a new SIV in pig can occur through virus transmission from another species, antigenic change in the major viral antigens through mutations (drift), or genetic reassortment (exchange of genes between two or more viruses). Antigenic shift is observed when HA and NA genes are swapped during reassortment.
Viruses of three different subtypes (H1N1, H3N2, H1N2) are currently circulating in swine throughout the world but their origin is different depending on the area and several genetic lineages may exist within a subtype. In Europe, the predominant H1N1 SIVs are entirely of avian origin, introduced from waterfowl to pigs in 1979 and referred to as ‘‘avian-like swine H1N1” (H1avN1). An H3N2 influenza virus was introduced in European pigs shortly after the Hong-Kong pandemic in 1968, but only became widespread after it reassorted with the H1avN1 virus, acquiring 6 internal genes from this latter. This “human-like reassortant swine H3N2” (H3N2) is now the dominant genotype of H3N2 virus in European pigs. In 1994 the H3N2 acquired the HA gene of a human H1N1 virus from the 1980s, resulting in the now predominant “human-like reassortant swine H1N2” (H1huN2). These three SIVs co-circulated for many years and new reassortant viruses between these enzootic viruses (or between SIVs and seasonal human viruses) have been detected since 2000. Antigenic drift occured through emergence of second generation reassortants such as rH1avN2 and rH1huN1, but such isolations were sporadic usually and only one such rH1avN2 virus has supplanted the common H1huN2 in a region. Besides reassortment events, viral evolution through antigenic drift was limited in European pigs last years, whatever the subtype. Thus, the situation was rather stable in European pig herds until the emergence in 2009 of the pandemic H1N1 virus (H1N1pdm) of swine origin.
Swine Influenza Viruses detected in European pigs in 2014

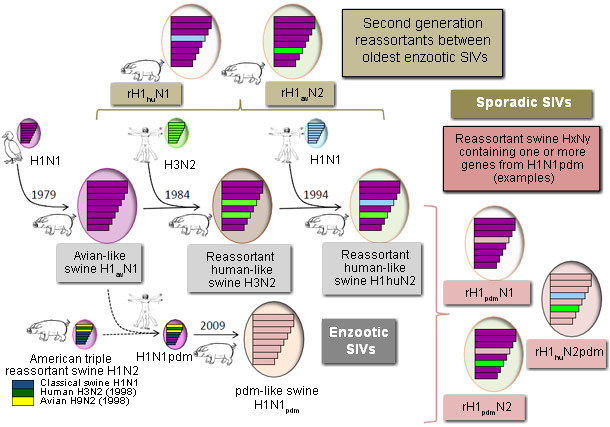
H1N1pdm has been generated from a reassortment between an American triple reassortant SIV (exhibiting a “classical” HA and genes of porcine, human and avian origins) and an H1avN1 virus from which the first acquired the NA and matrix (M) genes. Pigs were shown highly susceptible to this swine-origin human virus and numerous outbreaks of reverse zoonosis were reported worldwide very soon after its emergence in humans. Adaptation to swine and ongoing spread of the virus in pigs was then demonstrated in most main producing countries, including Europe. Soon after interspecies transmission, several reassortants between older enzootic SIVs and H1N1pdm were also detected. These novel “H1N1pdm-like reassortant viruses” consisted for instance in H1N1pdm viruses that acquired an N1 or N2 gene from European SIVs, or H1huN2 virus that acquired internal genes from H1N1pdm.
Thus, four virus lineages, having clearly distinguishable HA, are currently co-circulating in European pigs and can be considered as enzootic viruses, even if their relative prevalence and level of incidence vary from one country to another. The dominating lineage is still the oldest H1avN1, followed by the H1huN2, the H1N1pdm and the H3N2. The H3N2 subtype is missing from some geographic areas whereas it is still prevalent in others. Interestingly, areas free of H3N2 for some years are those that exhibit the highest frequencies of H1N2 (H1huN2 or rH1avN2) viruses. H1N2 reassortants containing genes from the H1N1pdm virus have been detected more and more frequently during the two last years, indicating that some of them might also become established in pig herds. They illustrate the increasing diversity and genetic complexity of European SIVs and potentially provide implications for zoonotic infections.
Continuing the surveillance and updating knowledge on the viruses, with shared contribution of farmers, veterinarians, scientists, stakeholders and authorities, is strongly encouraged.


