Along with Joan Lunney (ARS USDA) and Jack Dekkers (Iowa State University), the PRRS Host Genetics Consortium (PHGC) was established as a means to incorporate genomic and genetic tools to lessen the impact of PRRS. The approach combines state of the art methodologies in virology-immunology and genetics-genomics. The principal deliverables are the application of the results into commercial breeding programs. Previous attempts to identify genes that provide disease resistance have focused on comparing the disease response of different pure genetic lines of pigs. The PHGC does not make breed comparisons, but focuses on the identification of resistance/susceptibility markers that are present within populations of commercial animals exhibiting a diversity of disease phenotypes. The PHGC model incorporates the experimental infection of 200 young pigs with well-characterized PRRS viruses. Important phenotypic disease traits recorded for each pig include mortality, serum virus load, weight gain, neutralizing antibody and serum cytokine levels (see Figure 1 for examples). Genotyping all pigs with the porcine 60K SNP chip and conducting genome wide association studies have identified several markers linked with clinical disease progression and resistance. The favorable allele at one important marker on Sus scrofa chromosome 4 (SSC4) is associated with increased weight gain and decreased virus load. Over the course of the 42 day experimental infection period, pigs with the favorable genotype gained about 10% more weight and showed a corresponding decrease in viremia. The effect of the beneficial genotype is modeled in Figure 2. Results to date indicate that a similar benefit of the favorable allele can be achieved under farm conditions.

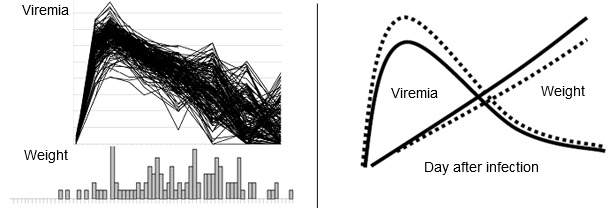
| Fig.1. PRRSV disease phenotypes at the population level. The above figures show PRRSV viremia (upper fig.) and weight gain distribution (lower fig.) in 200 pigs experimentally infected with PRRSV. Pigs were infected at 3 weeks of age and data collected for 42 days after infection. | Fig. 2. The favorable effect of the beneficial marker in SSC4 on weight gain and viremia in young pigs. The above model shows the expected improvements in weight gain and viremia in pigs with the favorable allele (AB or BB, solid line) versus pigs with the AA genotype (dashed lines). The increase in weight gain is approximately 10%. The decrease in peak viremia is approximately 1 log. |
Additional phenotypic outcomes are related to various aspects of PRRSV-specific immunity. For example, we identified an inverse relationship between virus load and virus neutralizing titer. In addition, a small percentage of infected pigs produce a unique antibody response, which can be described as broadly neutralizing; with the capacity to neutralize a wide range of isolates including type I viruses. Even though a heritable component has not been identified for the neutralizing response, the level of anti-N protein specific antibody has a significant heritable component, and is linked to a SNP on SSC7. A marker on SSC1 correlates with severe respiratory disease in pigs with PRDC. The nearest genomic marker is in the vicinity of an inflammatory cytokine receptor.
The results from work conducted by the PHGC illustrate important findings. First, clinical disease and outcome following PRRS infection has significant inherited components. Secondly, a relatively small number of genomic markers are associated with a given disease phenotype. This makes it possible to incorporate marker-assisted selection as a means to breed pigs with improved responses to infection. And finally, to date, all genomic markers related to clinical disease and immunity map to regions of the pig genome that contain immunologically relevant genes, including interferon-stimulated genes, MHC genes and cytokine response genes. In conclusion, genomics and genetics are new tools to improve animal health, especially for those infectious diseases that lack effective vaccines or treatments. When improved genetics is combined with improved vaccines and better nutrition, it will become possible to reverse many of the negative effects of endemic PRRS.