Escherichia coli is found naturally in the intestinal flora of pigs and, under certain circumstances, it causes a clinical picture of colibacillosis, a disease that can manifest itself as different conditions involving neonatal diarrhoea, post-weaning diarrhoea, oedema disease, septicaemia, etc...
Proper diagnosis includes a thorough anamnesis, a correct selection of samples and a complete differential diagnosis supported by various techniques. The mere detection of pathogenic strains does not justify the disease in every case due to the often finding of asymptomatic carriers. Identifying relevant virulence strains and differentiating them from other normal gut flora is a highly topical diagnostic challenge. And the real time PCR (qPCR) applied to the detection of virulence factors (VFs) of E. Coli has rised up as a powerful tool able to generate useful information.

Based on the extraction of nucleic acids from different biological matrices (culture, faeces, intestinal or rectal swab), qPCR assays recognize specific genomic regions of E. coli encoding different VFs. In this work, each trial was designed to detect a different VF (see Table 1). An additional qPCR (ECCO) was used to confirm the presence of E. coli in the samples and to ensure that all stages of the technique had been performed correctly. qPCR is a qualitative assay, but it's also quantitative (Figure 1), which is a significant advantage over its predecessor, conventional PCR.
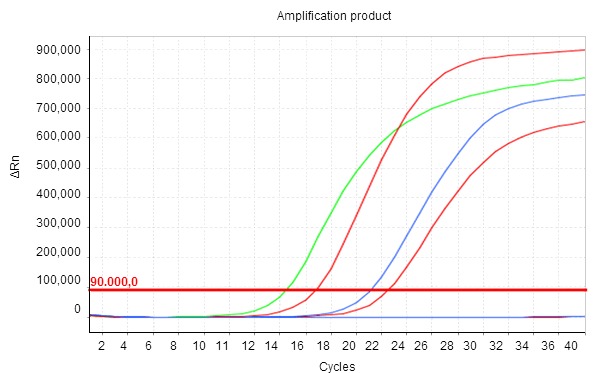
Figure 1. The amplification product is shown in real time as thermal cycles develop, so that the level of fluorescence recorded (ΔRn) increases exponentially. Cq is the number of cycles in which fluorescence begins to be detected. The smaller the Cq value, the higher the initial concentration of the parameter studied in the sample.
Evaluation of virulent strains in colibacillosis was traditionally achieved by an initial microbiological culture, then isolation of colonies of E. coli and subsequent characterization of their VFs. The main disadvantage of this method is that the analysis can only be performed on a limited number of selected isolates, the accuracy of the diagnosis relying on the assumption that these are representative in the total population of E. coli in the sample studied. This way, there is a risk of omitting non-majority populations of E. coli that are, however, clinically relevant.
qPCR provides the possibility of analysing the VFs directly on the clinical sample. This methodology avoids intermediate culture steps and their respective determinations on each of the different selected isolates, leading to considerable savings in time and costs. Given the quantitative nature of the technique, a relationship can be established between the number of copies detected for a specific virulence gene and the total population of E. coli in the sample. This way we could interpret the possibility of isolating a strain with a particular combination of VFs.
This methodology has certain limitations due to the indeterminate number of copies of the individual genes encoding each VF within different bacterial populations. However, it meets the proposed objective: to assess the possibility of finding E. coli with a particular combination of VFs in the sample.
Below is a practical example. Faced with a case of clinical suspicion of oedema disease in fattening pigs, four small intestines (SI) belonging all of them to clinical animals were sent to the laboratory to assess the presence of verotoxigenic E. coli. The pool, integrated by the 4 intestines, was directly evaluated by qPCR, with very similar Cq values for F18, Stx2e and ECCO (Table 1). These results can be interpreted as the presence of a large majority population of E. coli having the genes encoding for F18 and Stx2e, i.e., a verotoxigenic strain. A weakly positive value for F4 (Cq = 36.9) was also obtained, which would indicate a very minor presence of populations with that VF.
Simultaneously, a microbiological culture was performed with the samples, and 12 colonies of E. coli were selected. Phenotypic comparison revealed the presence of a single strain (Figure 2). Its characterization by determination of its VFs by qPCR confirmed that it was a potential verotoxigenic strain because genes encoding for F18 and Stx2e were detected.
Table 1. qPCR results.
| VF | Pool of 4 small intestines | E. coli strain isolated |
| F4 | POS (Cq = 36.9) | neg |
| F5 | neg | neg |
| F6 | neg | neg |
| F18 | POS (Cq = 22.28) | POS (Cq = 16.58) |
| F41 | neg | neg |
| LT | neg | neg |
| STa | neg | neg |
| STb | neg | neg |
| Stx2e | POS (Cq = 22.39) | POS (Cq = 17.29) |
| eae | neg | neg |
| ECCO | POS (Cq = 21.1) | POS (Cq = 16.97) |
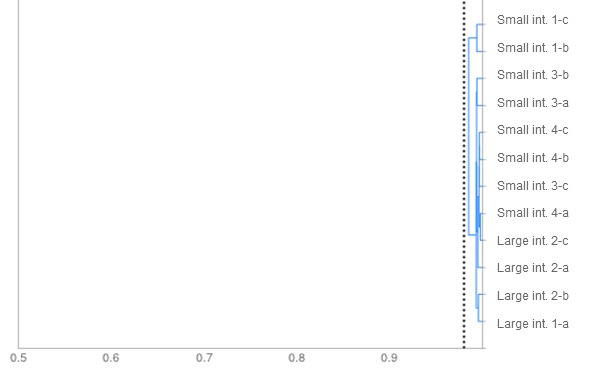

Figure 2. Dendrogram. The degree of similarity among isolates of E. coli according to their sugar fermentation pattern is shown. All isolates bear a similarity higher than the cut-off point, set at 0.98 (dotted line), which allows us to conclude that this is a unique strain.
In conclusion, qPCR applied directly to clinical samples provides substantial savings of time and resources, as well as information leading to an interpretation providing solutions for the sensitive diagnosis of porcine colibacillosis.



