Genetic improvement in the pig has been very successful over the past 50 years. The typical percentage of annual genetic progress, independent of all the improvements in feed, management, health and environment, has been:
| Numbers born alive per litter | +0.15 |
| Growth rate (grams per day) | +0.41 |
| Lean growth | +0.79 |
| Feed conversion | -0.69 |
| Backfat (mm) | -1.66 |
Source: Walters, 2011

This improvement has been achieved through using a range of techniques:
- Controlled mating programmes
- Introgression through AI
- Minimised inbreeding
- Accurate performance testing
- Within-breed selection
- Sire and dam line selection objectives
- Selection indices for specific markets
- Animal model Best Linear Unbiased Prediction (BLUP)
BLUP has been a particularly powerful tool for improvement as it allows evaluation of future breeding candidates based on genetic potential alone. In other words, it removes the effects of differences in health status, feeding system, building design, season, location, etc. It does this through sophisticated statistical models using an animal’s own performance plus that of all its relatives in the pedigree over a range of different years, farms and environments.
Fig. Chromosome to genes to nucleotides
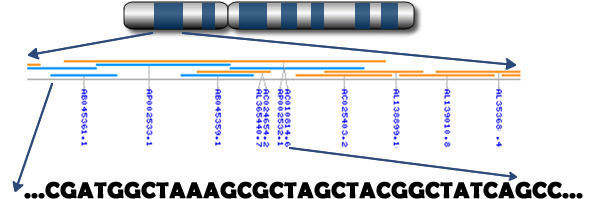
Genomics uses DNA information to improve the estimation of genetic merit
(Source: Prof. C.Haley – Roslin Institute, Edinburgh)
Over the last 20 years there has been much research on the use of DNA ‘markers’ to improve selection but results mostly have been disappointing. The main exceptions have been the success of selection for major genes such as halothane and E.coli resistance.
Now, following the publication of the draft porcine genome in 2009 and the recent publication of the ‘complete’ genome we are now into the potential new era of genomic selection. This follows the global dairy industry which is already using DNA information to select unproven bulls and cows rather than relying on expensive and less-accurate progeny testing.
Porcine SNP chip (Source: Illumina)
So what is genomic selection? Basically, it is the use of DNA information together with pedigree and performance data to predict a more accurate assessment of breeding value/genetic merit than ‘traditional’ BLUP. There are four main stages involved with genomic selection:
- DNA is sampled from individual blood, semen, hair follicle or tissue samples and then extracted in a laboratory.
- The DNA is analysed by means of a ‘gene chip’. This simultaneously measures differences in the genetic code between animals – often referred to as SNP’s (which stands for Single Nucleotide Polymorphism). These are single differences in the nucleotides or letters in the DNA. These SNP’s link specific segments of DNA to performance differences across the full range of measured and recorded traits.
- A DNA/SNP key (based on a prediction equation) is developed which works out the relation between the genes and the phenotypes. This has to be done in a large group of many thousands of animals (called the reference or training population) which is specific to a particular breed or population within a breed.
- The DNA/SNP key is used to translate all the available information (pedigree, testing, DNA) into a genomic breeding index.
What are the advantages of genomic selection?
The key benefits from the use of DNA information are:
- Greater overall genetic progress. For example, simulation studies in Denmark suggest an increase in genetic progress of 20-25% in dam lines and 10-15% in sire lines.
- Improved selection for ‘difficult’ traits, such as piglet survival, sow longevity and most meat and eating quality traits.
- Better control of inbreeding
Despite these advantages, there are several constraints with genomic selection:

- Despite the fact that the costs of genotyping have reduced spectacularly over the last few years, the overall cost of employing a genomic index are significantly higher than using a traditional BLUP index.
- Genotyping animals will never replace accurate performance recording in large numbers of animals. Thus, animals must still be tested and recorded – potentially for all traits of interest including those that are not currently included in breeding programmes.
- The DNA/SNP key needs constant recalibration as more animals come into a population and selection changes the ‘active’ genes in the population.
- Very high numbers of animals are required particularly for the reference populations. Like so much in animal breeding, it is a numbers game!
In the next article in the series the potential for improvements in pig health through genomic selection will be reviewed.



