In two recent articles ‘What do we mean by the genomics revolution?’ and ‘Genomics and pig health’ the science and potential of genomics was discussed. According to the Ensembl database, which stores data on the vertebrate genomes, the latest version of the available pig genome sequence has identified the following:
- 21,627 protein-coding genes.
- 3,024,658,701 base pairs.
- 484,949 Single Nucleotide Polymorphisms (SNP’s).

To date, genomics had very little impact on commercial production
The fact that there are close to half a million identified SNP’s means that at least some variations in sequence are already known for all the protein-coding genes. The task is now to harness this information to aid the drive to increased genetic progress and to ensure the successful integration of the SNP data with pedigree and performance data. This is being undertaken by publicly-funded research groups around the globe as well as by the major global pig breeding groups. So, what of progress to date? On the positive side, the key developments are, as follows:
- The reduction in cost of genotyping has been spectacular and easily matches the increase in computing power at lower and lower costs seen over the last 50 years.
- The introduction of new gene/SNP chips and the potential development of specific population chips.
- An increase in the number of traits being recorded accurately at the phenotype level in traits such as disease tolerance and resistance and meat and eating quality.
- Better understanding of the requirements for updating the genomic prediction equations in the reference population(s).
- The successful evolution of statistical models, such as GBLUP and single-step evaluation, to allow the integration of SNP data with the ‘traditional’ BLUP models which use pedigree and phenotype data (reproduction, performance, etc).
- The use of the statistical method of imputation which can convert low density marker genotypes to greater accuracy high density genotypes.
Genomics – future delivery may be important for health improvements.
As a result of these developments, some breeding companies are claiming significant genetic change. However, there is very little evidence of this in the scientific literature and several independent observers believe that commercial delivery has not yet been achieved. Of course, this may be for reasons of confidentiality but is more likely to be because of the constraints of genomic selection. These include:
- The requirement for large numbers of pigs in the reference populations to achieve robust accuracies for the genomic predictions – the associations between the SNP’s and performance. This aspect has very significant cost implications.
- Genomic predictions for animals in nucleus herds outside the reference population are only accurate if they are closely related to the reference population. As a result, genomic predictions decline in accuracy if the reference population is not fully maintained. Again, this is a significant cost factor.
- SNP’s linkages are breed and line specific. Although most pig breeding companies use Large White, Landrace and Duroc lines they are often quite distinct genetically and mostly many generations away from the lines of other breeding enterprises. Thus, it is not possible to transfer prediction equations from one line to another of the same breed or between breeds.
- As outlined in a previous article it is essential to have accurate performance recording in very large numbers of animals.
- The requirement for new breeding programme and data designs to capture the benefits of more accurate breeding values early in life (at birth or even in gestation).
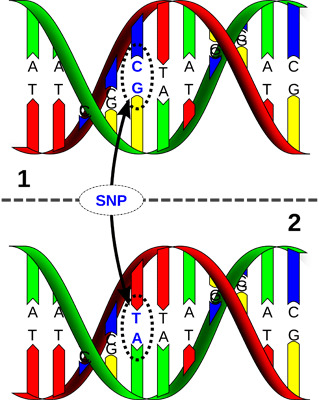
Genomics – linking SNP’s with performance. Source David Hall, via Wikimedia Commons
However, despite these constraints, some large companies are investing heavily in order to achieve the creation of company and product differentiation. This will allow delivery of some improvements from genomics in the next five years. However, it is likely that the optimal combination of conventional (BLUP) and genomic selection will still heavily favour the former, unless ‘new’ traits such as disease resistance/tolerance can be linked to specific SNP’s and then integrated successfully into populations.
Finally, does the complexity and expense of genomics mean that it will only be available to the major global pig breeding companies in the future? If the price of genotyping continues to reduce, it may be possible for medium and smaller size breeding companies to share funding and identify desired genotypes. However, this will require high levels of mutual cooperation.




